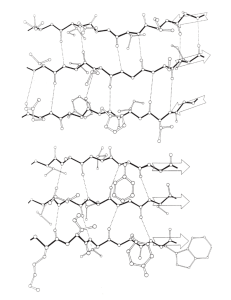
Try Pol X:DNA with 1
advertisement

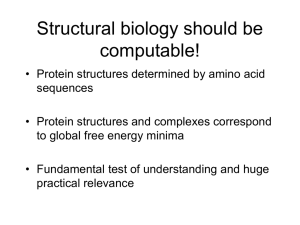
"Protein Design with the Molecular Modeling Software Rosetta" An overview will be given of Rosetta's protein design capabilities. These include methods for stabilizing proteins, increasing protein-protein binding affinities, design of novel protein interactions, de novo design of protein structure and enzyme design. Critical to many of these protocols is the simultaneous sampling of protein conformational space and sequence space. In this way it is possible search for conformations and docked positions that are intrinsically more designable. Rosetta methods for sampling conformational and sequence space will be discussed as well as the key features of its energy function.