The Cardiovascular Gene Ontology Initiative
advertisement

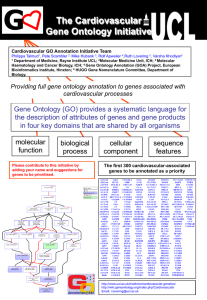
The Cardiovascular Gene Ontology Initiative Ruth Lovering1, Varsha Khodiyar1, Daniel Barrell2, Emily Dimmer2, Pete Scambler3, Mike Hubank4, Rolf Apweiler2, Philippa Talmud1 1Department of Medicine, Rayne Institute, UCL; 2Gene Ontology Annotation (GOA) Project, European Bioinformatics Institute, Hinxton; 3Molecular Medicine Unit, ICH; 4Molecular Haematology & Cancer Biology, ICH Providing full gene ontology annotation to genes associated with cardiovascular processes Gene Ontology (GO) provides a systematic language for the description of attributes of genes and gene products in three key domains molecular function biological process cellular component Call for annotation contributions We have provided a variety of tools to enable cardiovascular scientists to review the annotation of their “favourite” gene and suggest what information is missing, incomplete or inaccurate in these annotations. GO annotations can be viewed and commented on using the editable wiki site. Alternatively annotation suggestions can be sent by email or through the GOA user survey form. Useful URLs: Editable wiki site: http://wiki.geneontology.org/ index.php/Cardiovascular GOA user survey form: http://www.ebi.ac.uk/GOA/ contactus.html CV GOA Initiative home page: http://www.ucl.ac.uk/ medicine/cardiovascular-genetics/geneontology.html GOA home page: http://www.ebi.ac.uk/GOA/ Email: goannotation@ucl.ac.uk Figure showing part of the cardiac differentiation Gene Ontology, showing the different relationships between the parent and child terms. QuickGO enables terms of interest to be highlighted. The first 248 cardiovascular-associated genes to be annotated as a priority ABCA1 ABCG1 ACE ACE2 ADAM17 ADAM9 ADD1 ADIPOQ ADRA1A ADRA2A ADRA2B ADRB1 ADRB2 ADRBK1 AGT AGTR1 AGTR2 ANGPTL3 APOA1 APOA2 APOA4 APOA5 APOB APOC2 APOC3 APOE APOH APOL1 ATP2A2 AXIN1 BBS1 BBS2 BBS4 BBS5 BMP10 BMPR2 CALR CAPN10 CAV1 CAV3 CDH13 CDK6 CDKAL1 CDKN2A CDKN2A CDKN2AIP CDKN2B CDKN2C CDKN2D CELSR2 CETP CHD7 CHRD CHRNA1 CIDEA CILP2 CITED2 CLPTM1 COL2A1 CRKL CRP CST3 CXCL12 CXCR4 CYBA CYP11B1 CYP11B2 CYP26B1 CYP26C1 DRD1 DRD2 DRD3 DRD4 DVL1 ECE1 EDN1 EDN2 EGFR ELN ENG ENPP1 ERBB2 ERBB3 EVC EXT2 F12 F2R FGF8 FKBP1A FKBP1B FLNA FMN2 FOXC1 FOXJ1 FURIN FXN G6PD GAA GALNT2 GATA4 GATA6 GBX2 GHR GHRHR GJA1 GJA5 GNB3 GPX1 GYS1 HAS2 HBEGF HES1 HHEX HIC2 HIF1A HIRA HMOX1 HNF1A HNF1B HNF4A ICAM1 IFT80 IGF1 IGF2BP2 IL1B IL6R IL6ST INS INSR ISL1 JAG1 JMJD6 JUP KCNJ11 KDR KL KRIT1 LBP LDLR LEFTY2 LEP LEPR LPA LPL MAPK7 MEF2C MEN1 MIA3 MLXIPL MTHFD1L MTHFR MYBPC3 MYL1 MYL2 MYL3 MYLK2 MYLPF NEUROD1 NFATC1 NKX2-5 NKX2-6 NODAL NOG NOS1 NOS2A NOS3 NOX1 NPPA NPPB NR2F2 NRG1 OBSL1 P2RX4 P2RX7 PAFAH1B1 PBX4 PCK1 PCSK2 PCSK5 PCSK6 PDGFA PDGFC PDGFRA PITX2 PKD1 PLA2G1B PON1 PON3 PPARG PRKAG2 PRKAR1A PROX1 PSEN1 PSEN2 PSMD9 PSRC1 PTEN PTPN11 RARA RARB RARG RBP4 REN ROBO1 ROBO2 ROBO3 ROBO4 RPS19 RXRA SCD SELE SELS SEMA3C SERPINE1 SIRT1 SLC30A8 SLC6A4 SLIT1 SLIT2 SLIT3 SMAD3 SMAD4 SOD2 SOD2 SORT1 SOX4 SRF TBL2 TBX2 TCF7L2 TGFB2 TGFBR2 TGFBR3 TH THBS1 TMSB4X TNF TNNC1 TPM1 TRIB1 TTN USF1 VCAM1 VCL VEGFA VNN1 WFS1 WRN