Bringing Gene Ontology to cardiovascular research
advertisement

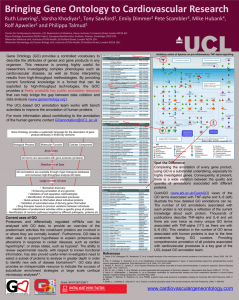
Bringing Gene Ontology to cardiovascular research Ruth Lovering1, Varsha Khodiyar1, Emily Dimmer2, Tony Sawford2, Peter Scambler3, Mike Hubank4, Rolf Apweiler2, Philippa J Talmud1 1 Centre for Cardiovascular Genetics, Department of Medicine, University College London, Rayne Institute, 5 2 University Street, London WC1E 6JF, UK, European Bioinformatics Institute, Hinxton, Cambridge, CB10 3 1SD, UK. Molecular Medicine Unit, Institute of Child Health, 30 Guilford Street, London WC1N 1EH, UK. 4 Molecular Hematology and Cancer Biology Unit, Institute of Child Health, 30 Guilford Street, London WC1N 1EH, UK Email: r.lovering@ucl.ac.uk, Tel: +44 20 7679 6965 URL: www.cardiovasculargeneontology.com Gene Ontology (GO) vocabularies are an established standard for linking functional information to genes and gene products (www.geneontology.org). A recent collaboration between University College London and the European Bioinformatics Institute is providing GO annotation to human cardiovascular-associated genes (www.cardiovasculargeneontology.com) The overall objective of the Cardiovascular Gene Ontology (GO) Annotation Initiative is to provide a unique public resource for the cardiovascular community both in the UK and internationally, by providing comprehensive functional annotation for genes implicated in heart development and cardiovascular processes and disease. High-throughput genomics and systems biology are particularly powerful tools for the investigation of multi-factorial phenotypes, such as cardiovascular disease. However, the interpretation of high-throughput analysis is often limited by the quality and quantity of the annotations available. Consequently, the ability to identify discriminatory groupings within a dataset can be vastly improved by additional annotations, associating more specific functions to the genes in the dataset. Comprehensive annotation of the human genome using GO is a formidable undertaking and although several approaches have been instigated to achieve this, more manual annotation is still required. In addition to annotating gene products associated with cardiovascular systems and disease the Cardiovascular Gene Ontology (GO) Annotation Initiative is helping to improve the ontology of GO, in areas such as heart development, apoptosis, signalling pathways and cardiac conduction (See Figure 1). This continued expansion of GO allows gene product annotators to comprehensively capture highly specific information relevant to genes involved in cardiovascular disease. The Cardiovascular GO Annotation Initiative aims to focus manual annotation on the human genes involved in cardiovascular-related processes and systems. This concentrated effort to improve the information content of the GOC dataset will facilitate effective interpretation of the data resulting from highthroughput methodologies and will be of benefit to many researchers seeking to alleviate cardiovascular diseases. This project is supported by the British Heart Foundation grant BHF SP/07/007/23671. Figure 1. QuickGO view of GO (www.ebi.ac.uk/QuickGO) displays 5 of the 280 heart development terms, the majority of which have been created through BHF funding.