pIns‐c‐MycER in vivo in silico Yi‐Fang Wang
advertisement
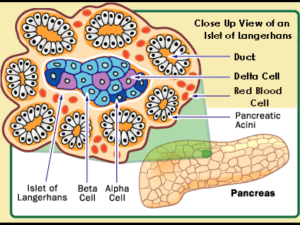
β‐cell turnover and insulin secretion in pIns‐c‐MycERTAM mice in vivo from laboratory data to in silico models Warwick Postgraduate Research Scholarship Yi‐Fang Wang1, Hugo van den Berg2,3, David Epstein2, Michael Khan1 1Department of Biological Sciences, 2Department of Mathematics, University of Warwick, 3Warwick Systems Biology Centre 3. Results 1. Introduction c-MycERTAM is a fusion protein which mediates the activation by 4-hydroxytamoxifen (4-OHT) of c-Myc in pancreatic β-cells in a transgenic mouse model (Pelengaris et al. 1999). Following 4-OHT-induced activation of c-myc, this oncogene stimulates both proliferation and apoptosis, with the latter dominating after a brief transitory period. Massive β-cell death leads to hyperglycaemia (high glucose). However, during the transition period, hypoglycaemia (low glucose) is observed, which may be caused by any combination of the following factors: reduced food intake; increased energy expenditure; increased insulin secretion; passive release of insulin from dying cells; rapid β-cell proliferation; increased insulin sensitivity. We aim to characterize the initial stages of the expression of c-myc in the plns-c-MycERTAM mouse model and to identify the causes of the transient hypoglycaemia by means of experiments and mathematical models. 2. Methods (1) plns‐c‐MycERTAM mouse model (5) Simulation from glucose, insulin and β‐cell mass dynamics (1) Hypoglycaemia starts within 16 hrs of c‐myc activation (A) (B) Glucose (mmol/l) Remote insulin (pmol/l) (C) Figure 5. Simulated time series of glucose (G(t)), plasma insulin (Ip(t)) and remote insulin (Ii(t)) for standard (reference) parameter values. There is a sustained oscillation. Figure 3. Glucose level of TL experiments (n=3). Glucose level of (A) WT, (B) Myc without treatment, and (C) Myc with treatment mice. Figure 2. Experimental design for glucose and energy expenditure dynamics in vivo. (3) Microarray analysis Total RNA was extracted from islets, which were isolated by laser capture microdissection from pancreas sections. Total RNA was processed the two-cycle target labeling reaction and hybridized to Affymetrix MOE430 Plus 2.0 GeneChip microarrays (Robson, 2008). Triplicate samples were analyzed at 4, 8, 16, 32 and 72 hours after c-Myc activation using Microarray Suite version 5.0 and normalized by the robust multiarray average (GC-RAM). Results were tested for statistical significance using 2-way ANOVA. Plasma insulin dynamics dI p Remote insulin dynamics Ip I dI i I = E( − i ) − i , dt V p Vi ti 2 dG = Gin − f 2 (G ) − f 3 (G ) f 4 ( I i ) + f 5 ( x3 ), dt 3 Time delays (2) Glucose and energy expenditure dynamics dt = Qf1 (G ) − E ( Ip Vp − Ip Ii )− , Vi tp Proliferation (r) Apopto1sis (d0) 4‐OHT Insulin secretion rate (Rm) 4‐OHT (3) Hypolycaemia results from hyperinsulinaemia (A) (B) Glucose (mmol/l) Glucose (mmol/l) Plasma insulin (pmol/l) Plasma insulin (pmol/l) Remote insulin (pmol/l) Remote insulin (pmol/l) (C) 1 (D) dQ G (G / V g ) = (−d 0 + r ( − ))Q, dt Vg Gc 4 dx1 3 = ( I p − x1 ), dt t d 5 dx2 3 = ( x1 − x2 ), dt td 6 dx3 3 = ( x 2 − x3 ), dt td 7 (E) (F) 4. Discussion & outlook Ip: Plasma insulin level; Q: Relative β-cell mass; E: Insulin transportation rate between two compartments; Vp: Distribution volume for insulin in plasma; Vi: Effective volume of the intercellular space; Ii: Intercellular insulin level; tp: Time constant for insulin in plasma; ti: Time constant for insulin in intercellular space; Gin: Glucose infusion rate; d0: β-cell mass death rate at zero glucose; r: Rate constant; Gc: Glucose saturation constant of the response function; td: Time constant; x1-x3: auxiliary parameters for time delay. f1(G): Glucose stimulated insulin secretion (GSIS) f3(G): Glucose-depend glucose utilization Figure 4. Glucose and insulin levels at specific time points(n=3). Glucose level of (A) WT, (B) Myc without treatment, and (C) Myc with treatment mice. Insulin level of (D) WT, (E) Myc without treatment, and (F) Myc with treatment. (4) Gene expression analysis f2(G): Insulin-independent glucose utilization f4(Ii) Insulin-dependent glucose utilization Food intake, faeces production and body weight were recorded every 4 hrs during the experimental period. f5(x3): Hepatic glucose production Post-mortem serum insulin (Ultrasensitive Mouse Insulin ELISA, Mercodia) and mouse pancreata were collected after c-myc activation of 8, 12, 16, 20, 28, 32, and 36 hrs. Scenario II: Increased β‐cell proliferation, apoptosis, and insulin secretion rate (2) Altered energetic balance does not completely account for hypoglycaemia 2 β‐cell mass dynamics Whole-body glucose was measured using the oxidase method (Roche Accuchek® glucose monitoring strips) at 0, 4, 8, 12, 16, 20, 24, 28, 32 and 36 hrs after the activation of c-myc (MO) in vivo. Controls included vehicle (peanut oil) treatment of plns-cMycERTAM mice (MF) and wildtype (WT) littermates. Each experiment was performed in triplicate. Scenario I: Increased β‐cell proliferation and apoptosis (4) Dynamics of glucose, insulin and β‐cell mass Glucose dynamics Figure 1. c‐MycERTAM transgenic model. HAT, histone acetyl transferases. Plasma insulin (pmol/l) (Tolic et al., 2000; Topp et al., 2000) a) 2716 genes exhibited a statistically detectable response over the whole time series to the treatment (p<0.05; 2‐way ANOVA). b) Selected changes in gene expression following Myc activation in vivo Function Gene symbol Cell cycle (G2/M) Cell cycle (G2/M) Cell cycle (G2/M) Cell cycle (G1/S) Cell cycly(G1/S) Cell cycle (G1/S) Cell cycle (G1/S) Cell cycle (G1/S) Cell cycle Cell cycle Exocytosis Exocytosis Exocytosis Exocytosis Exocytosis Exocytosis GSIS GSIS Insulin‐signaling Insulin‐signaling Insulin‐signaling Insulin‐signaling Insulin‐signaling Insulin‐signaling Insulin‐signaling Anti‐apoptosis Anti‐apoptosis Anti‐apoptosis DNA repair DNA repair DNA repair Ccna2 Ccnb1 Ccnb2 Ccne1 Ccne2 Ccnd1 Ccnd2 Cdkn1b Cdkn2d E2f1 Rims2 Pclo Cplx1 Syt1 Syt12 Napg Slc2a2 Gck Ins2 Ins1 Irs1 Irs2 Igf1 Raf1 Pdk1 Pdx1 Neurod1 Lgals3 Rad51 Pcna Brca1 Gene title cyclin A2 cyclin B1 cyclin B2 cyclin E1 cyclin E2 cyclin D1 cyclin D2 cyclin‐dependent kinase inhibitor 1B (p27KIP1) cyclin‐dependent kinase inhibitor 2D (p19, inhibits CDK4) E2F transcription factor 1 regulating synaptic membrane exocytosis 2 piccolo (presynaptic cytomatrix protein) complexin 1 synaptotagmin I synaptotagmin XII N‐ethylmaleimide sensitive fusion protein attachment protein gamma solute carrier family 2 (facilitated glucose transporter), member 2 glucokinase insulin II insulin I insulin receptor substrate 1 insulin receptor substrate 2 insulin‐like growth factor 1 v‐raf‐leukemia viral oncogene 1 pyruvate dehydrogenase kinase, isoenzyme 1 pancreatic and duodenal homeobox 1 neurogenic differentiation 1 lectin, galactose binding, soluble 3 RAD51 homolog (S. cerevisiae) proliferating cell nuclear antigen breast cancer 1 UniGene ID Mm.4189 Mm.260114 Mm.22592 Mm.16110 Mm.35867 Mm.273049 Mm.333406 Mm.2958 Mm.464272 Mm.18036 Mm.309296 Mm.146275 Mm.5195 Mm.289702 Mm.262270 Mm.292687 Mm.18443 Mm.220358 Mm.4946 Mm.46269 Mm.4952 Mm.407207 Mm.268521 Mm.184163 Mm.34411 Mm.389714 Mm.4636 Mm.248615 Mm.471596 Mm.7141 Mm.244975 FC at FC at FC at FC at FC at 4hr ‐1.08 1.10 ‐1.29 2.30 2.28 2.27 2.03 ‐1.50 ‐1.10 ‐1.20 1.09 1.04 1.07 ‐1.62 3.53 ‐2.47 ‐1.08 ‐1.40 ‐4.42 ‐3.82 1.24 ‐1.49 ‐1.02 ‐2.71 5.45 ‐1.62 ‐1.06 1.63 1.13 1.18 ‐1.06 8hr 2.37 1.92 ‐2.96 3.96 5.19 3.27 1.96 ‐2.17 ‐2.20 3.04 3.41 3.87 2.33 ‐2.38 4.55 ‐1.64 2.69 ‐2.69 ‐1.29 1.50 ‐1.16 ‐1.54 ‐1.10 ‐2.29 5.44 ‐1.99 4.90 ‐1.73 3.05 1.35 1.07 16hr 4.73 4.14 5.22 3.16 3.45 ‐1.46 1.30 ‐1.56 3.59 1.17 ‐1.89 ‐1.11 2.15 ‐1.87 1.26 1.18 ‐1.14 1.72 3.95 5.57 2.34 1.25 ‐1.43 2.27 4.23 ‐1.33 ‐1.03 15.31 4.34 5.09 3.74 32hr 3.83 2.84 1.18 1.92 5.18 1.19 1.54 ‐2.38 ‐1.56 ‐1.50 ‐1.36 ‐1.93 1.39 1.15 2.22 ‐1.19 ‐1.71 ‐2.30 11.25 10.72 1.47 ‐2.14 2.05 1.10 4.25 ‐3.47 1.08 8.37 1.79 2.96 1.69 72hr 1.78 3.25 2.38 1.05 1.88 1.74 1.25 1.36 1.48 1.17 1.58 1.97 1.58 1.02 1.04 1.63 1.28 1.32 1.62 1.66 1.16 1.20 1.60 1.21 1.91 1.23 1.54 8.86 1.09 1.99 1.27 Our results indicate that hypoglycaemia results from hyperinsulinaemia and is not due to passive release of insulin from dying β‐cells, as previously suggested by Cano et al. (2008): hyperinsulinaemia precedes the decrease in β‐cell mass in plns‐c‐MycERTAM mice and, moreover, hypoglycaemia still occurs when Myc‐induced apoptosis is fully prevented by co‐expression of the anti‐apoptotic protein BclxL in plns‐c‐ MycERTAM/RIP‐BclxL double‐transgenic mice (unpublished data). Microarray analysis implicates the signalling pathways of both the cell cycle and insulin secretion, although not yet conclusively. Myc‐triggered insulin release may be related to cell cycle entry and/or cell turn over. It is possible that autocrine or paracrine insulin release is a physiological regulator of β‐cell turn over. If this is the case, the massive synchronized insulin release from c‐myc activated cells might lead to “inadvertent” hypoglycaemia. To investigate this hypothesis, we will quantify β‐cell mass during the hypoglycaemic transient and also quantify proliferating and apoptotic cells during this period by means of immunohistochemistry. We will use the mathematical model to investigate whether the observed trends can explain the hypoglycaemia. Cano, D. A., Rulifson, I. C., Heiser, P. W., Swigart, L. B., Pelengaris, S., German, M., Evan, G. I., Bluestone, J. A. and Hebrok, M. (2008). Regulated beta-cell regeneration in the adult mouse pancreas. Diabetes. 57(4):958-966. Pelengaris, S., Khan, M. and Evan, G.I. (2002). Suppression of Myc-induced apoptosis in β−cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 109(3):321-334. Robson, S. C. (2008). Life or cell death: identifying c-Myc regulated genes in two distinct tissues. Ph. D. Thesis, Doctoral Training Centre for Molecular Organisation and Assembly in Cells, University of Warwick. Topp, B., Promislow, K., deVries, G., Miura, R. M. and Finegood, D. T. (2000). A model of β-cell mass, insulin, and glucose kinetics: pathways to diabetes. J. Theor. Biol. 206(4):605-619. Tolic, I. M., Mosekilde, E. and Sturis, J. (2000). Modeling the insulin-glucose feedback system: the significance of pulsatile insulin secretion. J. Theor. Biol. 207: 361-375. Contact: Yi-Fang.Wang@warwick.ac.uk