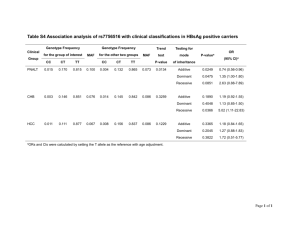
Power of a genome-wide association study (GWAS) for
advertisement

Power of a genome-wide association study (GWAS) for finding a gene for handedness Chris McManus, April 2012 [Note: This was a working document, and is provided only as an example of the approach to the calculations, and final checks on it have not been made. It was subsequently superseded by the Monte Carlo simulations]. The Twin Research Unit data on handedness in twins provides information on a large, if self-selected and hence non-representative group of twins, for whom handedness is known in many cases, and SNPs have also been mapped. Although the sample appears large, at over 2000 pairs, the proposed gene for handedness is not easy to map, having one phenotype which is random, and the heterozygote is also additive, both of which reduce power, and therefore leaving open the question of whether there is sufficient power, particularly if the GWAS has a negative outcome. 1) The McManus genetic model for handedness. The McManus model of the genetics of handedness has two unusual features, both of which seem to be essential for it successfully to fit the large amounts of family data and twin data in the literature: a) Although one of the alleles, D for Dextral, is determinate, in the sense that DD homozygotes are all right-handed, the other allele, C for Chance, is indeterminate, the CC genotype producing a 50:50 mixture of right and left handers. Although this in some ways looks like partial penetrance, the phenotype is better conceptualised as having 100% penetrance for fluctuating asymmetry (and the model as such is equivalent to those for situs inversus in mice and other species). That the randomness is not merely measurement error, or results from the effects of non-identified genetic variation1, means that it is better conceptualised as ‘deep chance’, in a fundamental sense. b) The heterozygote, DC, is neither dominant nor recessive, but instead is additive, 25% of DC individuals being left-handed (i.e. midway between the 0% for DD individuals and the 50% for CC individuals). There is no theoretical reason for the model being additive, but empirically the model fails to fit family and twin data when DC is not additive. 2) The six different genetic models of handedness. Table 1 compares six different types of genetic model for handedness, in order to assess the typical effects of sample size for detecting allelic association with a single SNP (a priori) and also with the approximately 2.5 million SNPs in a GWAS. Although only one of the six models actually fits the family and twin data, the other five are useful for assessing the effects on power of different components of the models. The six possible genetic models are: a) DC models. The two alleles are D and C (i.e. one of the alleles, C, produces fluctuating asymmetry when homozygous). Three variants are fitted in which DC is recessive, additive or dominant. 1 That is seen well in iv/iv mice, which carry the recessive iv gene which results in a 50:50 mixture of situs inversus (si) and situs solitus (ss). Crosses of known iv/iv mice, where the parental pairs are phenotypically ss x ss, ss x si, or si x si in each case result in 50% si in the offspring, implying there is no remaining genetic variance, and that fluctuating asymmetry is indeed the proper description of the phenotype. 1 b) DS models. These models, which have an S (Sinistral) allele, are more typical of Mendelian models, the SS homozygote producing left-handedness in all individuals. Once again three variants are fitted, with DS recessive, additive or dominant. DS dominant and DS recessive are typical of standard Mendelian recessive and dominant models. c) Allele frequencies. For the purposes of table 1, a) allele frequencies have been adjusted in each model so that p(L), the population proportion of left-handers is always 10%; b) the minor allele frequency of the SNP has been set equal to p(C) or p(S), in order that there can be maximal cis association between the D&C/D&S alleles and the A&a alleles of the SNP. 3) The calculations. The calculations follow, in several ways, the methods of Ohashi et al [1], in particular using their straightforward method for calculating the N required for a 2x2 contingency table, using proportions, rather than the cumbersome, and non-intuitive, noncentral chi-square. The results are also restricted to what Ohashi et al call an ‘allele frequency table’, in which there are two [independent] alleles from each individual. Note that as a result, N in the table applies to the number of participants, whereas those N participants contribute 2N alleles, and it is on the latter that the power is calculated. 4) Differences in power of different genetic models. As had long been suspected, Table 1 shows that different genetic models show different power. In general, recessive models are more powerful than dominant, which in turn are more powerful than additive (for both DC and DS models), and DS models are generally more powerful than DC models. The genetic model which is most likely to be correct in terms of fitting family and twin patterns of handedness, the DC additive model, has substantially less power for finding associations with SNPs than any of the other models. 5) Difficulties of interpretation of table 1. It must be emphasised that table 1 maximises the probability of an association being found by the fact that the MAF of the SNP is set to the same value as the frequency of the C allele. As a result there can be a perfect association in cis linkage disequilibrium between the C [or S allele] and the a allele (and the D allele with the A allele). That is generally not the case in a GWAS, MAFs taking all possible values between 0 and 0.5, making a real GWAS less powerful, as will be considered in the next section. 6) Restriction of remaining analyses to the standard DC additive mode. Since the most likely genetic model for handedness is DC additive, and table 1 has shown that it is the model with the least power for being detected, the remainder of the power analysis will restrict itself to the DC additive model, for which p(C)=0.2, 7) Power estimates in relation to the MAF of SNPs, and the distance between SNPs. The calculations of Table 1 were based entirely on the situation where the MAF is exactly the same as p(C) or p(S), the proportion of the minor allele in the genes underpinning handedness, which gives the maximum power. In reality, MAFs can take a range of values, and SNPs can be located at different distances from genes of interest, each of which can be considered separately. 8) Distribution of MAFs of SNPs. MAFs of SNPs can take values anywhere between zero and 0.5, in what, excluding those where the MAF is precisely zero (due to sampling error in a finite population), is close to a rectangular distribution. Power declines as the MAF becomes further and further removed from 0.2 (for examples see below in figure 1), essentially because, in a 2x2 contingency table, if the marginal proportions of rows and columns are different then the maximal number of cases on the diagonal (i.e. concordance between rows and columns) 2 cannot be 1, there having to be cases in the off-diagonal elements. The p(L) of 0.2 is close to the mean MAF of the SNPs, which is about 0.25 given the rectangular distribution, but nevertheless there are many SNPs with MAFs of, say, 0.01 or 0.49. 9) Distance of SNP from the handedness gene. Wheresoever the handedness gene is located, the nearest SNP will not be immediately adjacent, so that the SNP and the DC alleles, despite being in perfect linkage disequilibrium, are not perfectly correlated in a statistical sense. The association of a nearby SNP with the DC alleles will decrease as the recombination fraction (RF) increases, and that RF can also be expressed in terms of base-pairs, using the Haldane mapping function, which summarises the likely distance between a SNP and DC, given RF. 10) Power estimates for different MAFs and RFs. Using the same method as in Table 1, for any particular MAF and RF, but only with the DC additive model and p(C)=0.2, one can calculate Ncrit, the number of subjects needed in order to have a Bonferroni corrected alpha of 0.05/2,500,000 = 2x10-8, and beta = 0.99 (the latter being higher than the conventional 0.8 since one wishes to be strongly assured that an association has not slipped through the net). Lest the calculations are not clear, they are described on a worked, step-by-step basis in the appendix. a) Effect of MAF upon power. Figure 1 shows Ncrit (on the ordinate) in relation to RFs of 0 to 0.49, on the abscissa. The separate lines are, in the top row figures, for SNP MAFs of 0.001, 0.01, 0.02, 0.035, 0.5, 0.1 and 0.2, and in the bottom row figures, for SNP MAFs of 0.2, 0.25, 0.3, 0.35, 0.4, 0.45 and 0.5, the solid black line in each case being for MAF=0.2, which is also p(C). Power (i.e. lower values of Ncrit) is greatest for MAF=p(C)=0.2, and declines somewhat for MAFs > 0.2, and declines much more precipitously for MAFs < 0.2, with very low power as MAF approaches zero (and clearly there must be zero power for MAF = 0, when Ncrit = Infinity). b) Effect of RF. Power is also maximal for RF=0, and decreases as RF increases, and would necessarily be zero (Ncrit = Infinity) for RF=0.5. c) Relationship of power to distance in basepairs. The two right-hand figures in figure 1 replot the same results, except that RF has now been transformed, via the Haldane mapping function, into centimorgans, and then, on the basis of the usual assumption that 1 centimorgan = 1 million base-pairs (bp), into basepairs. Since the range is potentially large, the abscissa has actually been plotted as log10(base pairs) [although it is worth noting that if bp is on a linear scale then there is actually a straight line relationship of Ncrit to bp]. 11) The actual sample size for TRU. The TRU data, for present purposes, were regarded as two independent sets of singletons. Considering just set 1, and restricting the analysis to just chromosome 1, N for the various SNPs had a mean number of participants of 2257 (median = 2328, 95%iles 1937 and 2355). Given those values, a working estimate for the actual sample size of TRU of 2,200 has been used. The dashed, black horizontal line in the graphs of figure 1 is set at 2,200, and shows those combinations for which TRU has adequate power (below the line) and those for which it is underpowered. 12) The distances between the SNPs. Once again, Chromosome 1 has been used to assess the typical distribution of inter-SNP intervals measured in base-pairs. Of the 141,472 SNPs on chromosome 1, MAF=0 in 21,006 cases (14.8%), and they have therefore been removed from the analysis. Of the remaining 120,466 SNPs, the distribution of MAFs was close to rectangular, except for a strong peak at very low MAFs. Figure 1 suggests that MAFs below about 0.05 are rarely going to have sufficient power to find an association, and therefore MAFs below 0.05 were also removed from the analysis, leaving 89,392 viable SNPs. Of these SNPs, a few had low 3 Ns (e.g. the 1st percentile for N was 1225, and the 0.1th percentile was 304). In order to be within range of the typical N of 2,200, MAFs were also removed when N< 1800, leaving a final total of 85,801 SNPs. SNPs were then sorted into order by their position on the chromosome, and distances between adjacent SNPs calculated. The mean distance between SNPs was 1867 bp, but was very skewed, with a median of 725 bp, SD of 78,392 bp, and quartiles of 255 and 1809). Although 59.0% of gaps were <1 kbp, and 99% < 10kbp, there was a spattering of larger gaps, 12 between 100kbp and 199kbp, up to about 200 kbps, 5 between 200 and 999 kbps, one of 1.6 Mbps, and one of 22.8Mbps. The latter was between rs1851250 and rs861335, and quite where it comes from is very unclear. Since the 99th percentile of SNP gap length was 11,027 bp, and the 99.9th percentile was 32,413 bp, it was decided that if a SNP could detect an association within 30,000 bp (3 x 104 bp) then the genome would be reasonably covered. 13) Does the TRU data have adequate power for detecting SNP associations? The blue, dashed vertical lines in the right-hand side of figure 1 show the location of a gap of 30,000 bp (log10(30,000)=4.48) between a SNP and a putative handedness gene. It is clear that for associations less than 30,000 bps, and for all MAFs above 0.05 there is adequate power for detecting associations when N = 2200. The conclusion therefore has to be that the TRU database is adequately powered for detecting a DC additive genetic model for handedness. Reference List 1. Ohashi J, Yamamoto S, Tsichiya NHYKT, Matsushita M, Tokunaga K: Comparison of statistical power between 2x2 allele frequency and allele positivity tables in case-control studies of complex disease genes. AHG 2001, 65: 197-206. 4 Table 1: Comparison of different genetic models for handedness for a rate of left-handedness in the population, p(L), of 10%. The frequencies of the C or S allele differs between models in order to ensure p(L)=0.1. The DC additive model, highlighted in yellow, is considered to be the only reasonable fit to family and twin data on handedness. The minor allele frequency (MAF) of a single SNP with major and minor alleles A and a, is set to be the same as p(C) or p(S), thereby ensuring maximum power to detect association with cis linkage disequilibrium (LD). Linkage disequilibrium is set to be maximal (Recombination Frequency (RF) =0) or half of its maximal value (i.e. RF=.25). Of course when RF=.5 there is zero association. N crit, is the critical total number of individuals in the population to obtain a significant result with alpha = .05 and beta=.99 (i.e. a 99% chance of detecting an association if one is present) based on a population with 10% left-handers, and analyses either of a single SNP determined a priori (1SNP) or on the basis of a finding in a GWAS. Note that findings are not representative of a GWAS in general since it is not normally the case that p(a) = p(C) (or p(a)=p(S)). Genetic model DC recessive DC additive DC dominant DS recessive DS additive DS dominant p(L) for each genotype Gene frequency for p(L) = .10 MAF for SNP in cis Linkage Disequilibrium =p(a) Critical N for detecting association with a single SNP (alpha = .05, beta=.99) RF = 0 (i.e. perfect association) P(a) for SNP in left and right-handers P(a|L) P(a|R) .3858 1.0000 Ncrit DD DC CC p(C) p(a) 0 0 .5 .4472 .4472 0 .25 .5 .2000 .2000 .1556 .6000 0 .5 .5 .1056 .1056 .0587 .5279 16 / 43 41 / 183 27 / 145 DD DS SS p(S) p(a) P(a|L) P(a|R) Ncrit 0 0 1 .3162 .3162 .2402 1.0000 0 .5 1 .1000 .1000 .0500 .5500 0 1 1 .0513 .0513 .0000 .5132 1SNP / GWAS 10* / 28 23 / 125 15 / 102 Critical N for detecting association with a single SNP (alpha = .05, beta=.99) RF = 0 (i.e. perfect association) P(a) for SNP in left and right-handers P(a|L) P(a|R) .4165 .7236 N crit 1SNP / GWAS .1778 .4000 .0821 .3167 106 /397 166 / 721 106 / 516 P(a|L) P(a|R) Ncrit .2782 .6581 .0750 .3250 .0256 .2822 66 / 266 91 / 453 61 / 350 * If the value of 10 seems far too small to be possible, then consider a situation in which there is 1 left-hander and 9 right-handers. The left-hander under the DS recessive model with perfect linkage to the A/a alleles, must have haplotype SS/aa, while of the 9 right-handers, 4 will be DS/Aa and 5 will be DD/AA. Counting the 20 alleles, the left-hander(s) will have 2 a and 0 A, while the right-handers will have 4 a and 14 A. The 2x2 contingency table, ignoring corrections for small sample sizes etc, will have a chi-square= 5.19, which with 1 df has p=.023. While not worrying too much about the details of the calculation, the result is certainly order of magnitude correct. 5 Appendix: Example calculation for power for arbitrary MAF of SNP, and arbitrary RF, for DC additive model. Genetic model of handedness. For the DC additive model, p(L|DD)=0, p(L|DC=.25) and p(L|CC)=.5. For P(L) = .1, therefore p(C) = 2 x p(L) = .2, and p(D) = .8. SNP. The SNP has two alleles, A and a. Arbitrarily let p(a), the minor allele frequency (MAF) be .4, so that p(A)=1-p(A) = .6. Gamete haplotypes. The haplotype frequencies of the gametes can be considered firstly in the two extreme situations, of no association of D/C with A/a (i.e. recombinant frequency, RF = .5), and complete linkage disequilibrium (i.e. RF = 0) Random association of A/a with D/C (RF = 0.5) The entries in each cell of the contingency table reflect the marginal proportions D C A P(A) = .6 .6 x .8 = .48 .6 x .2 = .12 a .4 x .8 = .32 P(D)=.8 .4 x .2 = .08 P(C)=.2 P(a) = .4 1 cis linkage disequilibrum of A/a with D/C (RF = 0) The off-diagonal elements are as close to zero as possible. However, because the marginal probabilities are not equal, both off-diagonal elements cannot be zero D C A P(A) = .6 .60 .00 a .20 P(D)=.8 .20 P(C)=.2 P(a) = .4 1 For any arbitrary RF, say, 0.2, the gamete haplotypes can then be considered as a proportional mixture of the two extreme situations: A a RF = 0.2 RF = .2 can be considered as a mixture of 60% RF=0 and 40% RF=0.5. D C .48 x .4 + .60 x .6 .12 x .4 + .00 x.6 =.552 = .048 P(A) = .6 .32 x .4 + .20 x .6 .08 x .4 + .20 x .6 =.152 P(a) = .4 P(D)=.8 P(C)=.2 1 =.248 Given the frequencies of the four gametic haplotypes, it is then straightforward, using the HardyWeinberg equilibrium, to calculate the proportions of the sixteen haplotypes (i.e. DD/AA, DD/Aa, DD/aA, DD/aa, DC/AA, etc). 6 Haplotype DD/AA DD/aA DC/AA DC/Aa DD/Aa DD/aa DC/Aa DC/aa DC/AA DC/aA CC/AA CC/Aa DC/Aa DC/aa CC/Aa CC/aa Frequency 0.3047 0.1369 0.0265 0.0839 0.1369 0.0615 0.0119 0.0377 0.0265 0.0119 0.0023 0.0073 0.0839 0.0377 0.0073 0.0231 P(L|Haplotype) 0 0 0.25 0.25 0 0 0.25 0.25 0.25 0.25 0.5 0.5 0.25 0.25 0.5 0.5 Because the proportion of right- and left-handers in each haplotype is known (i.e. p(L|DD/xx)=0, p(L|DC/xx=.25) and p(L|CC/xx)=.5, where xx = any of AA, Aa, aA and aa), the frequencies of the SNP genotypes in right and left-handers can then be calculated: aa Right-handers Aa .4248 (47.2% .0552 (55.2%) .48 .1296 (14.4%) Left-handers .0304 (30.4%) .16 AA .3456 (38.4%) .0144 (14.4%) .36 .9 .1 1 And similarly the proportion of A and a alleles in the two phenotypes can be calculated. a Right-handers Left-handers .3420 (38.0%) .0580 (58.0%) .4 A .5580 (62.0%) .0420 (42.0%) .6 .9 .1 1 Given the proportions of a and A in the right-handers and left-handers in the table above, one can calculate the critical N required to have, say, a 99% probability (beta) of detecting those proportions, at a significance level of 0.05 (alpha), which may need Bonferroni correction for multiple testing. For a single test in a GWAS (alpha = 0.05/2,500,000, on the basis that there are 2.5 million SNPs, giving the -8 conventional significance level of 2x10 ), and using the formulae in Ohashi et al (2003), the critical N is 2066. However that N is for the number of alleles, and since each person in the study contributes two, independent alleles, the critical N in practice is 2066/2 = 1033 people. Conversion of RF to a distance in base pairs. The Haldane mapping function converts a recombination fraction, RF, to a distance, d, in Morgans, where d= -loge(1-2RF)/2. One Morgan is conventionally equivalent to 1million basepairs (bp), and hence there is a direct conversion. For RF = 0.2 in the example above, d = 0.255 7 Morgans = 25.5 centiMorgans = 25,500,000 basepairs = 2.55x10 bp. The Kosambi map function (d=loge((1+2.RF)/(1-2.RF))/4 ) could have been used as well, but in practice it makes little difference except at high levels of R 7 Figure 1 Plot of log10(Ncrit)for alpha=p<2x10 - 8, beta=.99 against log10(mBP) distance by MAF Standard DC model (pL|DD=0 pL|DC=.25 pL|CC=.5, pC=.2) 8 0.001 0.01 7 0.02 0.035 0.05 6 0.1 0.2 Plot of log10(Ncrit)for alpha=p<2x10 - 8, beta=.99 against RF by MAF Standard DC model (pL|DD=0 pL|DC=.25 pL|CC=.5, pC=.2) 8 7 log10(Ncrit) 6 log10(Ncrit) 0.001 0.01 0.02 0.035 0.05 0.1 0.2 5 4 4 3 3 2 2 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 Recombinant Fraction 0.4 0.45 0.5 Plot of log10(Ncrit)for alpha=p<2x10 - 8, beta=.99 against RF by MAF Standard DC model (pL|DD=0 pL|DC=.25 pL|CC=.5, pC=.2) 5.5 5 log10(Ncrit) 0.5 0.45 0.4 0.35 0.3 0.25 0.2 6 4.5 4 3.5 3 3 2 8 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 Recombinant Fraction 4.5 5 5.5 6 6.5 log10(BP) 7 7.5 8 8.5 4 3.5 2.5 4 Plot of log10(Ncrit)for alpha=p<2x10 - 8, beta=.99 against log10(mBP) distance by MAF Standard DC model (pL|DD=0 pL|DC=.25 pL|CC=.5, pC=.2) 6.5 0.5 6 0.45 0.4 5.5 0.35 0.3 5 0.25 0.2 4.5 6.5 log10(Ncrit) 5 2.5 0.4 0.45 0.5 2 4 4.5 5 5.5 6 6.5 log10(BP) 7 7.5 8 8.5