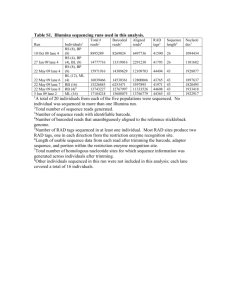
RAD Sequencing Design and Data Analysis John Davey RAD Sequencing Meeting
advertisement

RAD Sequencing Design and Data Analysis John Davey Institute of Evolutionary Biology University of Edinburgh RAD Sequencing Meeting Wednesday 21 October 2009 RAD Sequencing Design and Data Analysis Experimental Design John Davey Institute of Evolutionary Biology University of Edinburgh RAD Sequencing Meeting Wednesday 21 October 2009 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes John Davey Institute of Evolutionary Biology University of Edinburgh RAD Sequencing Meeting Wednesday 21 October 2009 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality John Davey Institute of Evolutionary Biology University of Edinburgh RAD Sequencing Meeting Wednesday 21 October 2009 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality John Davey Institute of Evolutionary Biology University of Edinburgh RAD Sequencing Meeting Wednesday 21 October 2009 SNP Discovery RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Illumina GAIIx Sequencing Machine RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane 8 lanes per run RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane 8 lanes per run 6 Gb per run RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane 8 lanes per run 6 Gb per run RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane 8 lanes per run 6 Gb per run RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane 8 lanes per run 6 Gb per run RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane 8 lanes per run 6 Gb per run RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane 8 lanes per run 6 Gb per run RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane 8 lanes per run 6 Gb per run RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane 8 lanes per run 6 Gb per run RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane 8 lanes per run 6 Gb per run RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane 8 lanes per run 6 Gb per run RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes How many individuals? Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane 8 lanes per run 6 Gb per run RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes How many individuals? How much coverage? Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane 8 lanes per run 6 Gb per run RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes How many individuals? How much coverage? Genome size (and GC content) Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane 8 lanes per run 6 Gb per run RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane 8 lanes per run 6 Gb per run RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Number of reads per lane Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane 8 lanes per run 6 Gb per run RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Number of reads per lane Estimated number of RAD tags Data Quality SNP Discovery Illumina GAIIx Sequencing Machine 15 million reads per lane 50 base pairs per read (soon to be 100) 750 Mb per lane 8 lanes per run 6 Gb per run RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality How many individuals? How much coverage? Plutella xylostella Genome size (and GC content) Restriction enzyme Number of reads per lane Estimated number of RAD tags 339 Mb SNP Discovery RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Number of reads per lane Estimated number of RAD tags Plutella xylostella Lymnaea stagnalis 339 Mb 1.2 Gb SNP Discovery RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Number of reads per lane Estimated number of RAD tags Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb Restriction enzyme Number of reads per lane Estimated number of RAD tags 15 million 15 million 15 million RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb Restriction enzyme Number of reads per lane Estimated number of RAD tags SbfI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 15 million 15 million 15 million RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb Restriction enzyme Number of reads per lane 15 million Estimated number of RAD tags SbfI EcoRI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 15 million 15 million RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb Restriction enzyme Number of reads per lane 15 million 15 million 15 million Estimated number of RAD tags SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb Restriction enzyme Number of reads per lane 15 million 15 million 15 million Estimated number of RAD tags SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb Restriction enzyme Average fragment size Symmetric? Number of reads per lane 15 million 15 million 15 million Estimated number of RAD tags SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI Average fragment size Symmetric? Number of reads per lane 15 million 15 million 15 million Estimated number of RAD tags SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI Average fragment size Symmetric? Number of reads per lane 15 million 15 million 15 million Estimated number of RAD tags SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.25 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI Average fragment size Symmetric? Number of reads per lane 15 million 15 million 15 million Estimated number of RAD tags SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Average fragment size Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI 1 / 0.258 Symmetric? Number of reads per lane 15 million 15 million 15 million Estimated number of RAD tags SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Average fragment size Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI 65 Kb Symmetric? Number of reads per lane 15 million 15 million 15 million Estimated number of RAD tags SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Average fragment size Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI 65 Kb Symmetric? Number of reads per lane Estimated number of RAD tags 15 million 15 million 15 million 339Mb / 65Kb SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Average fragment size Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI 65 Kb Symmetric? Number of reads per lane Estimated number of RAD tags 15 million 15 million 15 million 5,172 tags SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI Average fragment size 65 Kb Symmetric? Yes Number of reads per lane Estimated number of RAD tags 15 million 15 million 15 million 5,172 tags SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI Average fragment size 65 Kb Symmetric? Yes Number of reads per lane Estimated number of RAD tags 15 million 15 million 15 million 10,345 tags SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI Average fragment size 65 Kb Symmetric? Yes Number of reads per lane Estimated number of RAD tags 15 million 15 million 15 million 10,345 tags Number of reads per tag SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI Average fragment size 65 Kb Symmetric? Yes Number of reads per lane 15 million Estimated number of RAD tags 10,345 tags Number of reads per tag 15m / 10345 15 million 15 million SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI Average fragment size 65 Kb Symmetric? Yes Number of reads per lane Estimated number of RAD tags Number of reads per tag 15 million 15 million 15 million 10,345 tags 1,450 reads SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI Average fragment size 65 Kb Symmetric? Yes Number of reads per lane Estimated number of RAD tags Number of reads per tag Number of individuals per lane 15 million 15 million 15 million 10,345 tags 1,450 reads 1 SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI Average fragment size 65 Kb Symmetric? Yes Number of reads per lane Estimated number of RAD tags Number of reads per tag Number of individuals per lane 15 million 15 million 15 million 10,345 tags 1,450 reads 12 SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI Average fragment size 65 Kb Symmetric? Yes Number of reads per lane Estimated number of RAD tags Number of reads per tag Number of individuals per lane 15 million 15 million 15 million 10,345 tags 120 reads 12 SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI Average fragment size 65 Kb Symmetric? Yes Number of reads per lane Estimated number of RAD tags 15 million 15 million 15 million 10,345 tags Number of reads per tag 60 reads Number of individuals per lane 24 SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI SbfI SbfI Average fragment size 65 Kb 65 Kb 65 Kb Symmetric? Yes Yes Yes 15 million 15 million 15 million Genome size (and GC content) Restriction enzyme Number of reads per lane Estimated number of RAD tags 10,345 tags Number of reads per tag 60 reads Number of individuals per lane 24 SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb SbfI SbfI SbfI Average fragment size 65 Kb 65 Kb 65 Kb Symmetric? Yes Yes Yes 15 million 15 million 15 million Genome size (and GC content) Restriction enzyme Number of reads per lane Estimated number of RAD tags 10,345 tags 36,621 tags 103,759 tags Number of reads per tag 60 reads 17 reads Number of individuals per lane 24 24 6 reads 24 SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb EcoRI EcoRI EcoRI Yes Yes Yes 15 million 15 million 15 million Average fragment size Symmetric? Number of reads per lane Estimated number of RAD tags Number of reads per tag Number of individuals per lane SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb EcoRI EcoRI EcoRI Yes Yes Yes 15 million 15 million 15 million Average fragment size Symmetric? Number of reads per lane Estimated number of RAD tags Number of reads per tag Number of individuals per lane SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 p=0.256 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Average fragment size Symmetric? Number of reads per lane Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb EcoRI EcoRI EcoRI 4 Kb 4 Kb 4 Kb Yes Yes Yes 15 million 15 million 15 million Estimated number of RAD tags Number of reads per tag Number of individuals per lane SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 p=0.256 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Average fragment size Symmetric? Number of reads per lane Estimated number of RAD tags Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb EcoRI EcoRI EcoRI 4 Kb 4 Kb 4 Kb Yes Yes Yes 15 million 15 million 15 million 165,526 tags 585,936 tags 1,660,156 tags Number of reads per tag Number of individuals per lane SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 p=0.256 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Average fragment size Symmetric? Number of reads per lane Estimated number of RAD tags Number of reads per tag Number of individuals per lane Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb EcoRI EcoRI EcoRI 4 Kb 4 Kb 4 Kb Yes Yes Yes 15 million 15 million 15 million 165,526 tags 90 reads 1 585,936 tags 1,660,156 tags 25 reads 1 9 reads 1 SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 p=0.256 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb BbvCI BbvCI BbvCI 15 million 15 million Average fragment size Symmetric? Number of reads per lane 15 million Estimated number of RAD tags Number of reads per tag Number of individuals per lane SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 p=0.256 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb BbvCI BbvCI BbvCI 15 million 15 million Average fragment size Symmetric? Number of reads per lane 15 million Estimated number of RAD tags Number of reads per tag Number of individuals per lane SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 p=0.256 p=0.257 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Average fragment size Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb BbvCI BbvCI BbvCI 16 Kb 16 Kb 16 Kb 15 million 15 million 15 million Symmetric? Number of reads per lane Estimated number of RAD tags Number of reads per tag Number of individuals per lane SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 p=0.256 p=0.257 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Average fragment size Symmetric? Number of reads per lane Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb BbvCI BbvCI BbvCI 16 Kb 16 Kb 16 Kb No 15 million No No 15 million 15 million Estimated number of RAD tags Number of reads per tag Number of individuals per lane SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 p=0.256 p=0.257 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Average fragment size Symmetric? Number of reads per lane Estimated number of RAD tags Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb BbvCI BbvCI BbvCI 16 Kb 16 Kb 16 Kb No 15 million 20,690 tags No No 15 million 73,242 tags 15 million 207,519 tags Number of reads per tag Number of individuals per lane SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 p=0.256 p=0.257 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery How many individuals? How much coverage? Genome size (and GC content) Restriction enzyme Average fragment size Symmetric? Number of reads per lane Estimated number of RAD tags Plutella xylostella Lymnaea stagnalis H. sapiens 339 Mb 1.2 Gb 3.4 Gb BbvCI BbvCI BbvCI 16 Kb 16 Kb 16 Kb No 15 million 20,690 tags No No 15 million 73,242 tags 15 million 207,519 tags Number of reads per tag 60 reads 17 reads Number of individuals per lane 12 12 6 reads 12 SbfI EcoRI BbvCI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ 5’...GAATTC...3’ 3’...CTTAAG...5’ 5’...CCTCAGC...3’ 3’...GGAGTCG...5’ p=0.258 p=0.256 p=0.257 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality How many individuals? How much coverage? Plutella xylostella Genome size (and GC content) Restriction enzyme 339 Mb SbfI Average fragment size 65 Kb Symmetric? Yes Number of reads per lane Estimated number of RAD tags 15 million 10,345 tags Number of reads per tag 60 reads Number of individuals per lane 24 SbfI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ p=0.258 SNP Discovery RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality How many individuals? How much coverage? Plutella xylostella Genome size (and GC content) Restriction enzyme 339 Mb (40%) SbfI Average fragment size 65 Kb Symmetric? Yes Number of reads per lane Estimated number of RAD tags 15 million 10,345 tags Number of reads per tag 60 reads Number of individuals per lane 24 SbfI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ p=0.258 SNP Discovery RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality How many individuals? How much coverage? Plutella xylostella Genome size (and GC content) Restriction enzyme 339 Mb (40%) SbfI Average fragment size 65 Kb Symmetric? Yes Number of reads per lane Estimated number of RAD tags 15 million 10,345 tags Number of reads per tag 60 reads Number of individuals per lane 24 SbfI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ p=0.26 * 0.32 SNP Discovery RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality How many individuals? How much coverage? Plutella xylostella Genome size (and GC content) Restriction enzyme 339 Mb (40%) SbfI Average fragment size 87 Kb Symmetric? Yes Number of reads per lane Estimated number of RAD tags 15 million 10,345 tags Number of reads per tag 60 reads Number of individuals per lane 24 SbfI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ p=0.26 * 0.32 SNP Discovery RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality How many individuals? How much coverage? Plutella xylostella Genome size (and GC content) Restriction enzyme 339 Mb (40%) SbfI Average fragment size 87 Kb Symmetric? Yes Number of reads per lane Estimated number of RAD tags 15 million 3,905 tags Number of reads per tag 60 reads Number of individuals per lane 24 SbfI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ p=0.26 * 0.32 SNP Discovery RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality How many individuals? How much coverage? Plutella xylostella Genome size (and GC content) Restriction enzyme 339 Mb (40%) SbfI Average fragment size 87 Kb Symmetric? Yes Number of reads per lane Estimated number of RAD tags Number of reads per tag Number of individuals per lane SbfI 5’...CCTGCAGG...3’ 3’...GGACGTCC...5’ p=0.26 * 0.32 15 million 3,905 tags 160 reads 24 SNP Discovery RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 60 reads Symmetric? Yes Number of individuals per lane 24 Plutella xylostella 15 million RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 60 reads Symmetric? Yes Number of individuals per lane 24 Plutella xylostella 15 million RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 60 reads Symmetric? Yes Number of individuals per lane 24 Barcode SbfI site Plutella xylostella 15 million ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 60 reads Symmetric? Yes Number of individuals per lane 24 Barcode SbfI site Plutella xylostella 15 million ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 60 reads Symmetric? Yes Number of individuals per lane 24 Barcode SbfI site Plutella xylostella 15 million ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 60 reads Symmetric? Yes Number of individuals per lane 24 Barcode Plutella xylostella 15 million SbfI site ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG RAD Tags Genomic DNA % of All Reads with p>0.01 at position 50.0 37.5 25.0 12.5 0 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 Position RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 60 reads Symmetric? Yes Number of individuals per lane 24 Barcode Plutella xylostella 15 million SbfI site ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG RAD Tags Genomic DNA 15.00 % of All Reads 11.25 7.50 3.75 0 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 Number of errors in read RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 60 reads Symmetric? Yes Number of individuals per lane 24 Barcode Plutella xylostella 15 million SbfI site ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG F M C1 C2 C3 C4 C5 C6 C7 F M E C C8 = = = = C9 C10 E1 E2 Father Mother Experiment Control E3 E4 (R/R) (R/S) (R/R) (R/R or R/S) E5 E6 E7 E8 E9 E10 E11 E12 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 60 reads Symmetric? Yes Number of individuals per lane 24 Barcode Plutella xylostella 7.5 million SbfI site ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG F M C1 C2 C3 C4 C5 C6 C7 F M E C C8 = = = = C9 C10 E1 E2 Father Mother Experiment Control E3 E4 (R/R) (R/S) (R/R) (R/R or R/S) E5 E6 E7 E8 E9 E10 E11 E12 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Barcode Plutella xylostella 7.5 million SbfI site ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG F M C1 C2 C3 C4 C5 C6 C7 F M E C C8 = = = = C9 C10 E1 E2 Father Mother Experiment Control E3 E4 (R/R) (R/S) (R/R) (R/R or R/S) E5 E6 E7 E8 E9 E10 E11 E12 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Barcode Plutella xylostella 7.5 million SbfI site ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG F M C1 C2 C3 C4 C5 24 7 72 45 121 122 3 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 E12 90 157 141 211 108 25 77 107 89 124 180 147 36 15 117 155 140 F M E C = = = = Father Mother Experiment Control (R/R) (R/S) (R/R) (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Barcode Plutella xylostella 7.5 million SbfI site ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG F M C1 C2 C3 C4 C5 24 7 72 45 121 122 9 0 38 0 0 1 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 E12 3 90 157 141 211 108 25 77 107 89 124 180 147 36 15 117 155 140 1 51 49 F M E C 32 = = = = 25 27 Father Mother Experiment Control 0 0 0 35 (R/R) (R/S) (R/R) (R/R or R/S) 0 55 45 0 5 44 47 43 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Barcode Plutella xylostella 7.5 million SbfI site ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG F M C1 C2 C3 C4 C5 24 7 72 45 121 122 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 E12 3 90 157 141 211 108 25 77 107 89 124 180 147 36 15 117 155 140 9 0 38 0 0 1 1 51 49 32 25 27 0 0 0 35 0 55 45 0 5 44 47 43 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 F M E C = = = = Father Mother Experiment Control 0 (R/R) (R/S) (R/R) (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Barcode Plutella xylostella 7.5 million SbfI site ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG F M C1 C2 C3 C4 C5 24 7 72 45 121 122 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 E12 3 90 157 141 211 108 25 77 107 89 124 180 147 36 15 117 155 140 9 0 38 0 0 1 1 51 49 32 25 27 0 0 0 35 0 55 45 0 5 44 47 43 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 1 0 1 0 0 0 0 0 F M E C = = = = Father Mother Experiment Control (R/R) (R/S) (R/R) (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Plutella xylostella 7.5 million ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Plutella xylostella 7.5 million ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Tag length = 26 Father p<0.01 p<0.1 p<1 3,048 23,402 42,746 Mother 15,702 148,940 294,441 Control 1 22,739 173,717 315,764 Control 2 19,447 136,710 242,588 Control 3 63,508 189,824 284,603 Control 4 72,155 208,394 298,976 Control 5 3,081 8,435 12,113 Control 6 58,195 192,807 291,177 Control 7 123,558 352,517 522,155 Control 8 78,095 199,258 275,231 Control 9 100,405 224,210 302,329 64,353 154,814 216,053 Experiment 1 105,588 250,483 338,414 Experiment 2 112,224 282,414 399,781 Experiment 3 67,902 171,671 239,129 Experiment 4 30,581 77,581 107,889 Experiment 5 43,969 155,502 244,158 Experiment 6 57,167 169,644 255,064 Experiment 7 78,945 231,898 335,162 Experiment 8 68,111 211,327 316,014 Experiment 9 94,087 261,847 408,983 Experiment 10 19,954 229,096 337,275 Experiment 11 1,728 174,902 326,866 Experiment 12 79,528 13,078 24,111 Control 10 TOTAL 1,384,070 4,242,471 6,431,022 Plutella xylostella 7.5 million ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Tag length = 26 Father p<0.01 p<0.1 p<1 3,048 23,402 42,746 Mother 15,702 148,940 294,441 Control 1 22,739 173,717 315,764 Control 2 19,447 136,710 242,588 Control 3 63,508 189,824 284,603 Control 4 72,155 208,394 298,976 Control 5 3,081 8,435 12,113 Control 6 58,195 192,807 291,177 Control 7 123,558 352,517 522,155 Control 8 78,095 199,258 275,231 Control 9 100,405 224,210 302,329 64,353 154,814 216,053 Experiment 1 105,588 250,483 338,414 Experiment 2 112,224 282,414 399,781 Experiment 3 67,902 171,671 239,129 Experiment 4 30,581 77,581 107,889 Experiment 5 43,969 155,502 244,158 Experiment 6 57,167 169,644 255,064 Experiment 7 78,945 231,898 335,162 Experiment 8 68,111 211,327 316,014 Experiment 9 94,087 261,847 408,983 Experiment 10 19,954 229,096 337,275 Experiment 11 1,728 174,902 326,866 Experiment 12 79,528 13,078 24,111 Control 10 TOTAL 1,384,070 4,242,471 6,431,022 Plutella xylostella 7.5 million ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Tag length = 26 Father p<0.01 p<0.1 p<1 3,048 23,402 42,746 Mother 15,702 148,940 294,441 Control 1 22,739 173,717 315,764 Control 2 19,447 136,710 242,588 Control 3 63,508 189,824 284,603 Control 4 72,155 208,394 298,976 Control 5 3,081 8,435 12,113 Control 6 58,195 192,807 291,177 Control 7 123,558 352,517 522,155 Control 8 78,095 199,258 275,231 Control 9 100,405 224,210 302,329 64,353 154,814 216,053 Experiment 1 105,588 250,483 338,414 Experiment 2 112,224 282,414 399,781 Experiment 3 67,902 171,671 239,129 Experiment 4 30,581 77,581 107,889 Experiment 5 43,969 155,502 244,158 Experiment 6 57,167 169,644 255,064 Experiment 7 78,945 231,898 335,162 Experiment 8 68,111 211,327 316,014 Experiment 9 94,087 261,847 408,983 Experiment 10 19,954 229,096 337,275 Experiment 11 1,728 174,902 326,866 Experiment 12 79,528 13,078 24,111 Control 10 TOTAL 1,384,070 4,242,471 6,431,022 Plutella xylostella 7.5 million ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Tag length = 26 Father p<0.01 p<0.1 p<1 3,048 23,402 42,746 Mother 15,702 148,940 294,441 Control 1 22,739 173,717 315,764 Control 2 19,447 136,710 242,588 Control 3 63,508 189,824 284,603 Control 4 72,155 208,394 298,976 Control 5 3,081 8,435 12,113 Control 6 58,195 192,807 291,177 Control 7 123,558 352,517 522,155 Control 8 78,095 199,258 275,231 Control 9 100,405 224,210 302,329 64,353 154,814 216,053 Experiment 1 105,588 250,483 338,414 Experiment 2 112,224 282,414 399,781 Experiment 3 67,902 171,671 239,129 Experiment 4 30,581 77,581 107,889 Experiment 5 43,969 155,502 244,158 Experiment 6 57,167 169,644 255,064 Experiment 7 78,945 231,898 335,162 Experiment 8 68,111 211,327 316,014 Experiment 9 94,087 261,847 408,983 Experiment 10 79,528 229,096 337,275 Experiment 11 19,954 174,902 326,866 Experiment 12 1,728 13,078 24,111 Control 10 TOTAL 1,384,070 4,242,471 6,431,022 Plutella xylostella 7.5 million ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Tag length = 26 Father p<0.01 p<0.1 p<1 3,048 23,402 42,746 Mother 15,702 148,940 294,441 Control 1 22,739 173,717 315,764 Control 2 19,447 136,710 242,588 Control 3 63,508 189,824 284,603 Control 4 72,155 208,394 298,976 Control 5 3,081 8,435 12,113 Control 6 58,195 192,807 291,177 Control 7 123,558 352,517 522,155 Control 8 78,095 199,258 275,231 Control 9 100,405 224,210 302,329 64,353 154,814 216,053 Experiment 1 105,588 250,483 338,414 Experiment 2 112,224 282,414 399,781 Experiment 3 67,902 171,671 239,129 Experiment 4 30,581 77,581 107,889 Experiment 5 43,969 155,502 244,158 Experiment 6 57,167 169,644 255,064 Experiment 7 78,945 231,898 335,162 Experiment 8 68,111 211,327 316,014 Experiment 9 94,087 261,847 408,983 Experiment 10 79,528 229,096 337,275 Experiment 11 19,954 174,902 326,866 Experiment 12 1,728 13,078 24,111 Control 10 TOTAL 1,384,070 4,242,471 6,431,022 Plutella xylostella 7.5 million ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG Mother 294,441 / 10,345 = 28.4 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Tag length = 26 Father p<0.01 p<0.1 p<1 3,048 23,402 42,746 Mother 15,702 148,940 294,441 Control 1 22,739 173,717 315,764 Control 2 19,447 136,710 242,588 Control 3 63,508 189,824 284,603 Control 4 72,155 208,394 298,976 Control 5 3,081 8,435 12,113 Control 6 58,195 192,807 291,177 Control 7 123,558 352,517 522,155 Control 8 78,095 199,258 275,231 Control 9 100,405 224,210 302,329 64,353 154,814 216,053 Experiment 1 105,588 250,483 338,414 Experiment 2 112,224 282,414 399,781 Experiment 3 67,902 171,671 239,129 Experiment 4 30,581 77,581 107,889 Experiment 5 43,969 155,502 244,158 Experiment 6 57,167 169,644 255,064 Experiment 7 78,945 231,898 335,162 Experiment 8 68,111 211,327 316,014 Experiment 9 94,087 261,847 408,983 Experiment 10 79,528 229,096 337,275 Experiment 11 19,954 174,902 326,866 Experiment 12 1,728 13,078 24,111 Control 10 TOTAL 1,384,070 4,242,471 6,431,022 Plutella xylostella 7.5 million ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG 294,441 / 10,345 = 28.4 Mother 28.4 * 0.1 = 2.8 Threshold RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Tag length = 26 Father p<0.01 p<0.1 p<1 3,048 23,402 42,746 Mother 15,702 148,940 294,441 Control 1 22,739 173,717 315,764 Control 2 19,447 136,710 242,588 Control 3 63,508 189,824 284,603 Control 4 72,155 208,394 298,976 Control 5 3,081 8,435 12,113 Control 6 58,195 192,807 291,177 Control 7 123,558 352,517 522,155 Control 8 78,095 199,258 275,231 Control 9 100,405 224,210 302,329 64,353 154,814 216,053 Experiment 1 105,588 250,483 338,414 Experiment 2 112,224 282,414 399,781 Experiment 3 67,902 171,671 239,129 Experiment 4 30,581 77,581 107,889 Experiment 5 43,969 155,502 244,158 Experiment 6 57,167 169,644 255,064 Experiment 7 78,945 231,898 335,162 Experiment 8 68,111 211,327 316,014 Experiment 9 94,087 261,847 408,983 Experiment 10 79,528 229,096 337,275 Experiment 11 19,954 174,902 326,866 Experiment 12 1,728 13,078 24,111 Control 10 TOTAL 1,384,070 4,242,471 6,431,022 Plutella xylostella 7.5 million ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG 294,441 / 10,345 = 28.4 Mother 28.4 * 0.1 = 2.8 Threshold 42,746 / 10,345 = Father 4.1 * 0.1 = Threshold 4.1 0.4 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Tag length = 26 Father p<0.01 p<0.1 p<1 3,048 23,402 42,746 Mother 15,702 148,940 294,441 Control 1 22,739 173,717 315,764 Control 2 19,447 136,710 242,588 Control 3 63,508 189,824 284,603 Control 4 72,155 208,394 298,976 Control 5 3,081 8,435 12,113 Control 6 58,195 192,807 291,177 Control 7 123,558 352,517 522,155 Control 8 78,095 199,258 275,231 Control 9 100,405 224,210 302,329 64,353 154,814 216,053 Experiment 1 105,588 250,483 338,414 Experiment 2 112,224 282,414 399,781 Experiment 3 67,902 171,671 239,129 Experiment 4 30,581 77,581 107,889 Experiment 5 43,969 155,502 244,158 Experiment 6 57,167 169,644 255,064 Experiment 7 78,945 231,898 335,162 Experiment 8 68,111 211,327 316,014 Experiment 9 94,087 261,847 408,983 Experiment 10 79,528 229,096 337,275 Experiment 11 19,954 174,902 326,866 Experiment 12 1,728 13,078 24,111 Control 10 TOTAL 1,384,070 4,242,471 6,431,022 Plutella xylostella 7.5 million ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG 294,441 / 10,345 = 28.4 Mother 28.4 * 0.1 = 2.8 Threshold 42,746 / 10,345 = Father 4.1 * 0.1 = Threshold 4.1 0.4 With thresholding of p<0.1 reads: Reads 4,242,471 3,978,032 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Tag length = 26 Father p<0.01 p<0.1 p<1 3,048 23,402 42,746 Mother 15,702 148,940 294,441 Control 1 22,739 173,717 315,764 Control 2 19,447 136,710 242,588 Control 3 63,508 189,824 284,603 Control 4 72,155 208,394 298,976 Control 5 3,081 8,435 12,113 Control 6 58,195 192,807 291,177 Control 7 123,558 352,517 522,155 Control 8 78,095 199,258 275,231 Control 9 100,405 224,210 302,329 64,353 154,814 216,053 Experiment 1 105,588 250,483 338,414 Experiment 2 112,224 282,414 399,781 Experiment 3 67,902 171,671 239,129 Experiment 4 30,581 77,581 107,889 Experiment 5 43,969 155,502 244,158 Experiment 6 57,167 169,644 255,064 Experiment 7 78,945 231,898 335,162 Experiment 8 68,111 211,327 316,014 Experiment 9 94,087 261,847 408,983 Experiment 10 79,528 229,096 337,275 Experiment 11 19,954 174,902 326,866 Experiment 12 1,728 13,078 24,111 Control 10 TOTAL 1,384,070 4,242,471 6,431,022 Plutella xylostella 7.5 million ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG 294,441 / 10,345 = 28.4 Mother 28.4 * 0.1 = 2.8 Threshold 42,746 / 10,345 = Father 4.1 * 0.1 = Threshold 4.1 0.4 With thresholding of p<0.1 reads: Reads Tags 4,242,471 3,978,032 126,004 15,541 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Tag length = 26 Father p<0.01 p<0.1 p<1 3,048 23,402 42,746 Mother 15,702 148,940 294,441 Control 1 22,739 173,717 315,764 Control 2 19,447 136,710 242,588 Control 3 63,508 189,824 284,603 Control 4 72,155 208,394 298,976 Control 5 3,081 8,435 12,113 Control 6 58,195 192,807 291,177 Control 7 123,558 352,517 522,155 Control 8 78,095 199,258 275,231 Control 9 100,405 224,210 302,329 64,353 154,814 216,053 Experiment 1 105,588 250,483 338,414 Experiment 2 112,224 282,414 399,781 Experiment 3 67,902 171,671 239,129 Experiment 4 30,581 77,581 107,889 Experiment 5 43,969 155,502 244,158 Experiment 6 57,167 169,644 255,064 Experiment 7 78,945 231,898 335,162 Experiment 8 68,111 211,327 316,014 Experiment 9 94,087 261,847 408,983 Experiment 10 79,528 229,096 337,275 Experiment 11 19,954 174,902 326,866 Experiment 12 1,728 13,078 24,111 Control 10 TOTAL 1,384,070 4,242,471 6,431,022 Plutella xylostella 7.5 million ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG 294,441 / 10,345 = 28.4 Mother 28.4 * 0.1 = 2.8 Threshold 42,746 / 10,345 = Father 4.1 * 0.1 = Threshold 4.1 0.4 With thresholding of p<0.1 reads: Reads Tags 4,242,471 3,978,032 126,004 15,541 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Tag length = 26 Father p<0.01 p<0.1 p<1 3,048 23,402 42,746 Mother 15,702 148,940 294,441 Control 1 22,739 173,717 315,764 Control 2 19,447 136,710 242,588 Control 3 63,508 189,824 284,603 Control 4 72,155 208,394 298,976 Control 5 3,081 8,435 12,113 Control 6 58,195 192,807 291,177 Control 7 123,558 352,517 522,155 Control 8 78,095 199,258 275,231 Control 9 100,405 224,210 302,329 64,353 154,814 216,053 Experiment 1 105,588 250,483 338,414 Experiment 2 112,224 282,414 399,781 Experiment 3 67,902 171,671 239,129 Experiment 4 30,581 77,581 107,889 Experiment 5 43,969 155,502 244,158 Experiment 6 57,167 169,644 255,064 Experiment 7 78,945 231,898 335,162 Experiment 8 68,111 211,327 316,014 Experiment 9 94,087 261,847 408,983 Experiment 10 79,528 229,096 337,275 Experiment 11 19,954 174,902 326,866 Experiment 12 1,728 13,078 24,111 Control 10 TOTAL 1,384,070 4,242,471 6,431,022 Plutella xylostella 7.5 million ACGTATGCAGGGTAGTGTTGTGCCTTTTAGCATGTGCCTTTTAGCATGTG 294,441 / 10,345 = 28.4 Mother 28.4 * 0.1 = 2.8 Threshold 42,746 / 10,345 = Father 4.1 * 0.1 = Threshold 4.1 0.4 With thresholding of p<0.1 reads: Reads Tags 4,242,471 3,978,032 126,004 15,541 RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length All reads Father 26 bp Unique tags 23,402 3,389 Mother 148,940 7,097 Control 1 173,717 6,410 Control 2 136,710 5,960 Control 3 189,824 6,836 Control 4 208,394 6,318 Control 6 192,807 6,886 Control 7 352,517 6,874 Control 8 199,258 6,817 Control 9 224,210 6,484 Control 10 154,814 6,453 Experiment 1 250,483 6,949 Experiment 2 282,414 7,167 Experiment 3 171,671 6,632 Experiment 4 77,581 5,987 Experiment 5 155,502 6,507 Experiment 6 169,644 6,698 Experiment 7 231,898 6,737 Experiment 8 211,327 6,355 Experiment 9 261,847 6,724 Experiment 10 229,096 6,452 Experiment 11 174,902 6,578 TOTAL 4,220,958 Plutella xylostella 7.5 million RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length All reads Father 26 bp Unique tags 23,402 3,389 Mother 148,940 7,097 Control 1 173,717 6,410 Control 2 136,710 5,960 Control 3 189,824 6,836 Control 4 208,394 6,318 Control 6 192,807 6,886 Control 7 352,517 6,874 Control 8 199,258 6,817 Control 9 224,210 6,484 Control 10 154,814 6,453 Experiment 1 250,483 6,949 Experiment 2 282,414 7,167 Experiment 3 171,671 6,632 Experiment 4 77,581 5,987 Experiment 5 155,502 6,507 Experiment 6 169,644 6,698 Experiment 7 231,898 6,737 Experiment 8 211,327 6,355 Experiment 9 261,847 6,724 Experiment 10 229,096 6,452 Experiment 11 174,902 6,578 TOTAL 4,220,958 Plutella xylostella SbfI site number variation with GC content 7.5 million RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length All reads Father 26 bp Unique tags 23,402 3,389 Mother 148,940 7,097 Control 1 173,717 6,410 Control 2 136,710 5,960 Control 3 189,824 6,836 Control 4 208,394 6,318 Control 6 192,807 6,886 Control 7 352,517 6,874 Control 8 199,258 6,817 Control 9 224,210 6,484 Control 10 154,814 6,453 Experiment 1 250,483 6,949 Experiment 2 282,414 7,167 Experiment 3 171,671 6,632 Experiment 4 77,581 5,987 Experiment 5 155,502 6,507 Experiment 6 169,644 6,698 Experiment 7 231,898 6,737 Experiment 8 211,327 6,355 Experiment 9 261,847 6,724 Experiment 10 229,096 6,452 Experiment 11 174,902 6,578 TOTAL 4,220,958 Plutella xylostella SbfI site number variation with GC content 50%: 10,345 7.5 million RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length All reads Father 26 bp Unique tags 23,402 3,389 Mother 148,940 7,097 Control 1 173,717 6,410 Control 2 136,710 5,960 Control 3 189,824 6,836 Control 4 208,394 6,318 Control 6 192,807 6,886 Control 7 352,517 6,874 Control 8 199,258 6,817 Control 9 224,210 6,484 Control 10 154,814 6,453 Experiment 1 250,483 6,949 Experiment 2 282,414 7,167 Experiment 3 171,671 6,632 Experiment 4 77,581 5,987 Experiment 5 155,502 6,507 Experiment 6 169,644 6,698 Experiment 7 231,898 6,737 Experiment 8 211,327 6,355 Experiment 9 261,847 6,724 Experiment 10 229,096 6,452 Experiment 11 174,902 6,578 TOTAL 4,220,958 Plutella xylostella SbfI site number variation with GC content 50%: 10,345 45%: 6,652 40%: 3,905 7.5 million RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length All reads Father 26 bp Unique tags 23,402 3,389 Mother 148,940 7,097 Control 1 173,717 6,410 Control 2 136,710 5,960 Control 3 189,824 6,836 Control 4 208,394 6,318 Control 6 192,807 6,886 Control 7 352,517 6,874 Control 8 199,258 6,817 Control 9 224,210 6,484 Control 10 154,814 6,453 Experiment 1 250,483 6,949 Experiment 2 282,414 7,167 Experiment 3 171,671 6,632 Experiment 4 77,581 5,987 Experiment 5 155,502 6,507 Experiment 6 169,644 6,698 Experiment 7 231,898 6,737 Experiment 8 211,327 6,355 Experiment 9 261,847 6,724 Experiment 10 229,096 6,452 Experiment 11 174,902 6,578 TOTAL 4,220,958 Plutella xylostella 7.5 million SbfI site number variation with GC content 50%: 10,345 45%: 6,652 40%: 3,905 From 42 454 contigs with SbfI sites RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length All reads Father 26 bp Unique tags 23,402 3,389 Mother 148,940 7,097 Control 1 173,717 6,410 Control 2 136,710 5,960 Control 3 189,824 6,836 Control 4 208,394 6,318 Control 6 192,807 6,886 Control 7 352,517 6,874 Control 8 199,258 6,817 Control 9 224,210 6,484 Control 10 154,814 6,453 Experiment 1 250,483 6,949 Experiment 2 282,414 7,167 Experiment 3 171,671 6,632 Experiment 4 77,581 5,987 Experiment 5 155,502 6,507 Experiment 6 169,644 6,698 Experiment 7 231,898 6,737 Experiment 8 211,327 6,355 Experiment 9 261,847 6,724 Experiment 10 229,096 6,452 Experiment 11 174,902 6,578 TOTAL 4,220,958 Plutella xylostella 7.5 million SbfI site number variation with GC content From 42 454 contigs with SbfI sites 50%: 10,345 45%: 6,652 40%: 3,905 28 tag pairs found RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length All reads Father 26 bp Unique tags 23,402 3,389 Mother 148,940 7,097 Control 1 173,717 6,410 Control 2 136,710 5,960 Control 3 189,824 6,836 Control 4 208,394 6,318 Control 6 192,807 6,886 Control 7 352,517 6,874 Control 8 199,258 6,817 Control 9 224,210 6,484 Control 10 154,814 6,453 Experiment 1 250,483 6,949 Experiment 2 282,414 7,167 Experiment 3 171,671 6,632 Experiment 4 77,581 5,987 Experiment 5 155,502 6,507 Experiment 6 169,644 6,698 Experiment 7 231,898 6,737 Experiment 8 211,327 6,355 Experiment 9 261,847 6,724 Experiment 10 229,096 6,452 Experiment 11 174,902 6,578 TOTAL 4,220,958 Plutella xylostella 7.5 million SbfI site number variation with GC content From 42 454 contigs with SbfI sites 50%: 10,345 45%: 6,652 40%: 3,905 28 tag pairs found 11 singletons found RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length All reads Father 26 bp Unique tags 23,402 3,389 Mother 148,940 7,097 Control 1 173,717 6,410 Control 2 136,710 5,960 Control 3 189,824 6,836 Control 4 208,394 6,318 Control 6 192,807 6,886 Control 7 352,517 6,874 Control 8 199,258 6,817 Control 9 224,210 6,484 Control 10 154,814 6,453 Experiment 1 250,483 6,949 Experiment 2 282,414 7,167 Experiment 3 171,671 6,632 Experiment 4 77,581 5,987 Experiment 5 155,502 6,507 Experiment 6 169,644 6,698 Experiment 7 231,898 6,737 Experiment 8 211,327 6,355 Experiment 9 261,847 6,724 Experiment 10 229,096 6,452 Experiment 11 174,902 6,578 TOTAL 4,220,958 Plutella xylostella 7.5 million SbfI site number variation with GC content From 42 454 contigs with SbfI sites 50%: 10,345 45%: 6,652 40%: 3,905 28 tag pairs found 11 singletons found 3 tag pairs not found RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F 26 bp Plutella xylostella 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 F = Father (R/R) M = Mother (R/S) E = Experiment (R/R) C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F 26 bp Plutella xylostella 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 ● F = Father (R/R) - M = Mother (R/S) - - - - E = Experiment (R/R) - - - - - - C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F 26 bp Plutella xylostella 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 Tags ● 602 F = Father (R/R) - M = Mother (R/S) - - - - E = Experiment (R/R) - - - - - - C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F - Plutella xylostella 26 bp 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 Tags ● 5 - F = Father (R/R) - - - - - - M = Mother (R/S) - - - - - - - - - - - - - 602 - - - - - - - - - - - 440 E = Experiment (R/R) C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F Plutella xylostella 26 bp 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 Tags ● - - - - - - - - - - - 602 - 5 - - - - - - - - - - - - - - - - - - - - 440 - 5 * * * * * * * * * - - - - - - - - - - - 100 F = Father (R/R) M = Mother (R/S) E = Experiment (R/R) C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F Plutella xylostella 26 bp 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 Tags ● - - - - - - - - - - - 602 - 5 - - - - - - - - - - - - - - - - - - - - 440 - 5 * * * * * * * * * - - - - - - - - - - - 100 - 19 18 - - - - - 45 41 - - - - - - - - - - - - 62 F = Father (R/R) M = Mother (R/S) E = Experiment (R/R) C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F Plutella xylostella 26 bp 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 Tags ● - - - - - - - - - - - 602 - 5 - - - - - - - - - - - - - - - - - - - - 440 - 5 * * * * * * * * * - - - - - - - - - - - 100 - 19 18 - - - - - 45 41 - - - - - - - - - - - - 62 62 / F = Father (R/R) M = Mother (R/S) 15,441 = 0.4% E = Experiment (R/R) C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F Plutella xylostella 26 bp 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 Tags ● - - - - - - - - - - - 602 - 5 - - - - - - - - - - - - - - - - - - - - 440 - 5 * * * * * * * * * - - - - - - - - - - - 100 - 19 18 - - - - - 45 41 - - - - - - - - - - - - 62 62 / 15,441 = 0.4% 222 = 4,194,304 F = Father (R/R) M = Mother (R/S) E = Experiment (R/R) C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F Plutella xylostella 26 bp 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 Tags ● - - - - - - - - - - - 602 - 5 - - - - - - - - - - - - - - - - - - - - 440 - 5 * * * * * * * * * - - - - - - - - - - - 100 - 19 18 - - - - - 45 41 - - - - - - - - - - - - 62 62 / 15,441 = 0.4% 222 = 4,194,304 1 / 4,194,301 = 0.00002% F = Father (R/R) M = Mother (R/S) E = Experiment (R/R) C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F Plutella xylostella 26 bp 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 Tags ● - - - - - - - - - - - 602 - 5 - - - - - - - - - - - - - - - - - - - - 440 - 5 * * * * * * * * * - - - - - - - - - - - 100 - 19 18 - - - - - 45 41 - - - - - - - - - - - - 62 62 / 15,441 = 0.4% 222 = 4,194,304 1 / 4,194,301 = 0.00002% 1 / F = Father (R/R) M = Mother (R/S) 15,441 = 0.006% E = Experiment (R/R) C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F Plutella xylostella 26 bp 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 Tags ● - - - - - - - - - - - 602 - 5 - - - - - - - - - - - - - - - - - - - - 440 - 5 * * * * * * * * * - - - - - - - - - - - 100 - 19 18 - - - - - 45 41 - - - - - - - - - - - - 62 62 / 15,441 = 0.4% 222 = 4,194,304 1 / 4,194,301 = 0.00002% 1 / 15,441 = 0.006% Most common ‘interesting’ segmentation pattern (of 6408 segmentation patterns) F = Father (R/R) M = Mother (R/S) E = Experiment (R/R) C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F Plutella xylostella 26 bp 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 Tags ● - - - - - - - - - - - 602 - 5 - - - - - - - - - - - - - - - - - - - - 440 - 5 * * * * * * * * * - - - - - - - - - - - 100 - 19 18 - - - - - 45 41 - - - - - - - - - - - - 62 62 / 15,441 = 0.4% 222 = 4,194,304 1 / 4,194,301 = 0.00002% 1 / 15,441 = 0.006% Most common ‘interesting’ segmentation pattern (of 6408 segmentation patterns) F = Father (R/R) M = Mother (R/S) E = Experiment (R/R) C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F Plutella xylostella 26 bp 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 Tags ● - - - - - - - - - - - 602 - 5 - - - - - - - - - - - - - - - - - - - - 440 - 5 * * * * * * * * * - - - - - - - - - - - 100 - 19 18 - - - - - 45 41 - - - - - - - - - - - - 62 F = Father (R/R) M = Mother (R/S) E = Experiment (R/R) C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F 26 bp Plutella xylostella 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 - 19 18 F = Father (R/R) - - - - - 45 41 M = Mother (R/S) - - - - - - E = Experiment (R/R) - - - - - - Tags 62 C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F 26 bp Plutella xylostella 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 - 19 18 - - - - - 45 41 - - - - - - - - - - - - Tags 62 36 of 62 tags have variants 1bp apart F = Father (R/R) M = Mother (R/S) E = Experiment (R/R) C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F Plutella xylostella 26 bp 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 - 19 18 - - - - - 45 41 - - - - - - Tags - - - - - - 62 - - - - - - 7 36 of 62 tags have variants 1bp apart - 5 - F = Father (R/R) - - - 3 - - M = Mother (R/S) - - - - - - - E = Experiment (R/R) C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F Plutella xylostella 26 bp 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 - 19 18 - - - - - 45 41 - - - - - - Tags - - - - - - 62 - - - - - - 7 11 24 40 64 65 91 93 96 57 98 84 71 11 36 of 62 tags have variants 1bp apart - 5 - - - - 3 - - - 6 43 47 43 75 62 78 24 35 74 F = Father (R/R) M = Mother (R/S) - - - - - - E = Experiment (R/R) C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F Plutella xylostella 26 bp 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 - 19 18 - - - - - 45 41 - - - - - - Tags - - - - - - 62 - - - - - - 7 6 43 47 43 75 62 78 24 35 74 11 24 40 64 65 91 93 96 57 98 84 71 11 * 26 22 42 37 23 17 15 41 29 7 36 of 62 tags have variants 1bp apart - 5 - - - - 3 - - 26 43 16 22 61 F = Father (R/R) - - M = Mother (R/S) - - - - - - - - E = Experiment (R/R) 8 29 35 32 C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F Plutella xylostella 26 bp 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 - 19 18 - - - - - 45 41 - - - - - - Tags - - - - - - 62 - - - - - - 7 6 43 47 43 75 62 78 24 35 74 11 24 40 64 65 91 93 96 57 98 84 71 11 * 26 - 26 43 16 22 61 - - 22 42 37 23 17 15 41 29 7 4 - - 13 54 - - - - - - 3 10 - 9 - 31 49 15 - 26 29 35 20 2 36 of 62 tags have variants 1bp apart - 5 - F = Father (R/R) - - - - 3 - - 19 45 - 26 - - M = Mother (R/S) - - - 9 - - - - - 11 12 22 - 17 - - 8 29 35 32 - 32 - 19 E = Experiment (R/R) - C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F Plutella xylostella 26 bp 7.5 million M C1 C2 C3 C4 C6 C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 - 19 18 - - - - - 45 41 - - - - - - Tags - - - - - - 62 - - - - - - 7 6 43 47 43 75 62 78 24 35 74 11 24 40 64 65 91 93 96 57 98 84 71 11 * 26 - 26 43 16 22 61 - - 22 42 37 23 17 15 41 29 7 4 - - 13 54 - - - - - - 3 10 - 9 - 31 49 15 - 26 29 35 20 2 36 of 62 tags have variants 1bp apart - 5 - - - - - 3 - - 19 45 - 26 - - - - - 9 - - - - - 11 12 22 - 17 - - 8 29 35 32 - 32 - 19 - 6 F = Father (R/R) M = Mother (R/S) E = Experiment (R/R) C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length F = Father (R/R) 26 bp M = Mother (R/S) Plutella xylostella E = Experiment (R/R) 7.5 million C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length 26 bp F Plutella xylostella M C1 C2 C3 C4 C6 CTACACGCTGAAAGACCCATATTCGA - 22 28 - - - CTACACGCTGAAAGACCCATGTTCGA - 26 - 26 43 16 22 CTACACGCTGAAAGACCCATTTTCGA 13 - 29 16 20 22 20 F = Father (R/R) M = Mother (R/S) 7.5 million C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 - 19 19 61 - 31 15 20 - - - - - - - - - 22 42 37 23 17 15 8 29 35 32 20 35 53 20 8 22 15 29 19 19 E = Experiment (R/R) 41 33 29 9 C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length 26 bp F Plutella xylostella M C1 C2 C3 C4 C6 CTACACGCTGAAAGACCCATATTCGA - 22 28 - - - CTACACGCTGAAAGACCCATGTTCGA - 26 - 26 43 16 22 CTACACGCTGAAAGACCCATTTTCGA 13 - 29 16 20 22 20 F = Father (R/R) M = Mother (R/S) 7.5 million C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 - 19 19 61 - 31 15 20 - - - - - - - - - 22 42 37 23 17 15 8 29 35 32 20 35 53 20 8 22 15 29 19 19 E = Experiment (R/R) 41 33 29 9 C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length 26 bp F Plutella xylostella M C1 C2 C3 C4 C6 7.5 million C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 CTACACGCTGAAAGACCCATATTCGA - 22 28 - - - CTACACGCTGAAAGACCCATGTTCGA - 26 - 26 43 16 22 CTACACGCTGAAAGACCCATTTTCGA 13 - 29 16 20 22 20 - 19 19 61 - 31 15 20 - - - - - - - - - 22 42 37 23 17 15 8 29 35 32 20 35 53 20 8 22 15 29 19 19 41 33 29 9 ATATCAGTGATCTTCCAAGTGCGATC ATATCAGTGATCTTCCAAGTGCGGTC ATATCAGTAATCTTCCAAGTGCGATC - 12 21 - - - - 13 - 16 47 47 32 3 - 36 19 43 37 51 - 39 44 64 - 73 38 40 - - - - - - - - - 34 53 46 34 15 33 47 56 57 38 22 43 44 35 33 38 31 36 36 39 54 29 34 27 GCGCCCCGCGCTTTGTCCGTGTGTAA GCGCCCCGCGCTTTGTCCGTGTGTAG - 4 - 11 - 16 24 39 - - - - - 16 34 32 15 29 30 F = Father (R/R) 9 - - - - 16 21 12 16 M = Mother (R/S) E = Experiment (R/R) - - 9 22 20 - - 8 19 48 C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length 26 bp F Plutella xylostella M C1 C2 C3 C4 C6 7.5 million C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 CTACACGCTGAAAGACCCATATTCGA - 22 28 - - - CTACACGCTGAAAGACCCATGTTCGA - 26 - 26 43 16 22 CTACACGCTGAAAGACCCATTTTCGA 13 - 29 16 20 22 20 - 19 19 61 - 31 15 20 - - - - - - - - - 22 42 37 23 17 15 8 29 35 32 20 35 53 20 8 22 15 29 19 19 41 33 29 9 ATATCAGTGATCTTCCAAGTGCGATC ATATCAGTGATCTTCCAAGTGCGGTC ATATCAGTAATCTTCCAAGTGCGATC - 12 21 - - - - 13 - 16 47 47 32 3 - 36 19 43 37 51 - 39 44 64 - 73 38 40 - - - - - - - - - 34 53 46 34 15 33 47 56 57 38 22 43 44 35 33 38 31 36 36 39 54 29 34 27 GCGCCCCGCGCTTTGTCCGTGTGTAA GCGCCCCGCGCTTTGTCCGTGTGTAG - 4 - 11 - 16 24 39 - - - - - 16 34 32 15 29 30 F = Father (R/R) 9 - - - - 16 21 12 16 M = Mother (R/S) E = Experiment (R/R) - - 9 22 20 - - 8 19 48 C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length 26 bp F Plutella xylostella M C1 C2 C3 C4 C6 7.5 million C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 CTACACGCTGAAAGACCCATATTCGA - 22 28 - - - CTACACGCTGAAAGACCCATGTTCGA - 26 - 26 43 16 22 CTACACGCTGAAAGACCCATTTTCGA 13 - 29 16 20 22 20 - 19 19 61 - 31 15 20 - - - - - - - - - 22 42 37 23 17 15 8 29 35 32 20 35 53 20 8 22 15 29 19 19 41 33 29 9 ATATCAGTGATCTTCCAAGTGCGATC ATATCAGTGATCTTCCAAGTGCGGTC ATATCAGTAATCTTCCAAGTGCGATC - 12 21 - - - - 13 - 16 47 47 32 3 - 36 19 43 37 51 - 39 44 64 - 73 38 40 - - - - - - - - - 34 53 46 34 15 33 47 56 57 38 22 43 44 35 33 38 31 36 36 39 54 29 34 27 GCGCCCCGCGCTTTGTCCGTGTGTAA GCGCCCCGCGCTTTGTCCGTGTGTAG - 4 - 11 - 16 24 39 - - - - - 16 34 32 15 29 30 F = Father (R/R) 9 - - - - 16 21 12 16 M = Mother (R/S) E = Experiment (R/R) - - 9 22 20 - - 8 19 48 C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery Genome size (and GC content) 339 Mb (40%) Number of reads per lane Restriction enzyme SbfI Estimated number of RAD tags 10,345 tags Average fragment size 65 Kb Number of reads per tag 30 reads Symmetric? Yes Number of individuals per lane 24 Read error threshold p<0.1 Read trim length 26 bp F Plutella xylostella M C1 C2 C3 C4 C6 7.5 million C7 C8 C9 C10 E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 CTACACGCTGAAAGACCCATATTCGA - 22 28 - - - CTACACGCTGAAAGACCCATGTTCGA - 26 - 26 43 16 22 CTACACGCTGAAAGACCCATTTTCGA 13 - 29 16 20 22 20 - 19 19 61 - 31 15 20 - - - - - - - - - 22 42 37 23 17 15 8 29 35 32 20 35 53 20 8 22 15 29 19 19 41 33 29 9 ATATCAGTGATCTTCCAAGTGCGATC ATATCAGTGATCTTCCAAGTGCGGTC ATATCAGTAATCTTCCAAGTGCGATC - 12 21 - - - - 13 - 16 47 47 32 3 - 36 19 43 37 51 - 39 44 64 - 73 38 40 - - - - - - - - - 34 53 46 34 15 33 47 56 57 38 22 43 44 35 33 38 31 36 36 39 54 29 34 27 GCGCCCCGCGCTTTGTCCGTGTGTAA GCGCCCCGCGCTTTGTCCGTGTGTAG - 4 - 11 9 - - - - 16 21 12 16 - 16 24 39 - - - - - 16 34 32 15 - - 8 19 48 29 30 CTTATAGGGACATGCTGGTTAAGGCT CTTATAGGGACATGCTGGTGAAGGCT - 13 4 19 6 - - - - 32 42 30 36 103 4 10 - - - - - - - - - - - 20 77 62 47 18 32 34 26 71 40 99 22 TAGACCAGATGTCTGATGAATGGTGA TAGACCAGATGTCTGATGACTGGTGA - 8 17 - - - 3 4 13 - 31 54 31 46 - 17 27 91 - - - - - - - - - - - 17 70 46 55 19 50 58 68 44 37 77 32 TCATAATGGGCTCTTTTCCACCCACT TCATAATGGGCTCTTTTCCACCTACT - 14 20 - - - - 15 22 31 17 14 41 - 18 21 40 - - - - - 32 58 52 59 52 29 F = Father (R/R) M = Mother (R/S) E = Experiment (R/R) - - 9 22 20 - - - - - 6 35 50 33 15 26 C = Control (R/R or R/S) RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Data Quality SNP Discovery RAD Sequencing Design and Data Analysis Experimental Design Restriction Enzymes Mark Blaxter Marian Thomson Urmi Trivedi Karim Gharbi Simon Baxter Maureen Liu Eric Johnson Paul Etter Data Quality SNP Discovery