UNIVERSITY OF MALTA
advertisement

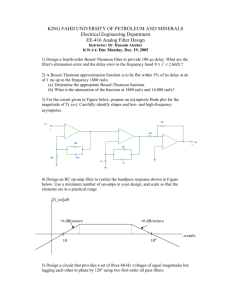
UNIVERSITY OF MALTA LIFE SCIENCE RESEARCH SEMINARS Web: http://www.um.edu.mt/events/scisem/ Email: scisem@um.edu.mt Abstract form Title: Restriction-site Associated DNA sequencing Presenter: Alexia Cardona Contact address: Laboratory of Molecular Genetics, Department of Physiology and Biochemistry, University of Malta Tel: 23403048 Fax: Email: alexia.cardona@um.edu.mt Presentation date: 2 May 2011 Abstract The Restriction-site Associate DNA (RAD) sequencing1 method is a genetic mapping method that is used in the study of population genetics. RAD sequencing utilises the Illumina Genome Analyser as a platform. The main idea of RAD sequencing is to digest the genome with restriction enzymes and sequence the regions surrounding restriction enzymes. Digesting genomes of different individuals with the same restriction enzyme will lead to the detection of SNPs across the genome as one can compare the different RAD tags of the different individuals and note the differences between them. The research done involves three main outcomes. First, the implementation of a tool that will generate a list of restriction enzyme fragments and RAD mappings that contain information such as the actual DNA sequence of the fragment, location of the fragment on the genome and other relevant information. The second outcome of this project is the analysis of the restriction enzyme fragments and RAD tags to be able to understand the data and to discover patterns in the data. The analysis done eventually leads to the third outcome of this project, the characterisation of fragment distribution of already sequenced genomes such as that of the nematode C. elegans. References 1.. Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA and Johnson EA. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE. 2008: 3, e3376.