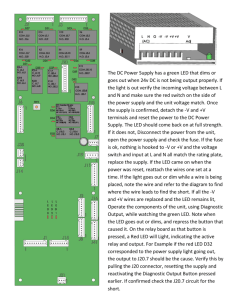
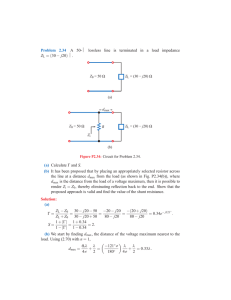
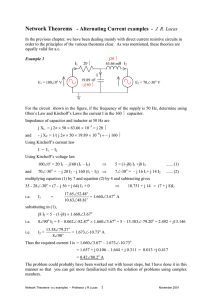
!"#$%&'()*+,-(."-/$,00#1",23-( 9&/5#:%"-(#;(9<=(9#>?0%(@%0&A( C:%"(DEF(#;(G'#H'(!"#$%&'(I$">5$>"%/(;"#J()*",-(
advertisement

BIOCH 590: Biomacromolecules
Spring 2010
!"#$%&'()*+,-(."-/$,00#1",23-(
Copyright: Jianhan Chen
456(7&,'3,'(.3%'(
9&/5#:%"-(#;(9<=(9#>?0%(@%0&A(
Watson, J. D., & Crick, F. H. C. A structure for
deoxyribose nucleic acid. Nature 171, 737–738 (1953)
456(7&,'3,'(.3%'(
8(
C:%"(DEF(#;(G'#H'(!"#$%&'(I$">5$>"%/(;"#J()*",-(
Discovery of DNA Structure and
Function: Watson and Crick, L. A.
Pray, Nature Education 1 (2008).
B(
First X-ray protein structures
Hemoglobin and myoglobin, by
Max Perutz and Sir John Kendrew,
respectively, in 1958
Source: PDB
456(7&,'3,'(.3%'(
K(
@&13*$3"#>132>$(2"#$%&'(5"-/$,00#1",23-N(
!"#$%&%'()*+)',-+./$$*0/1"&)
!"#$%&'(I$">5$>"%(9%$%"J&',L#'(?-()*",-(
."-/$,00#1",23-(
• !"#$%&'(2>"&M5,L#'N(M"/$(/$%2(#;(,00(/$">5$>",0(O%$%"J&',L#'(%P#"$/(
• !"#$%&'(."-/$,00&Q,L#'N($3%(J#/$(53,00%'1&'1(2,"$R(
– !"#O>5%(H%00*#"O%"%O(2"#$%&'(J#'#*5"-/$,0/(H&$3#>$(,'-(&'50>/&#'(,'O(0,"1%(
%'#>13($#(O&P",5$()*+,-(?%,J((
– S#"%(,"$($3,'(/5&%'5%N(0,"1%0-(&J2#//&?0%($#(2"%O&5$(5"-/$,00&Q,L#'(5#'O&L#'/(
• ."-/$,0(J#>'L'1N($",'/;%"(#;(5"-/$,0/(;"#J(/#0>L#'($#(O%$%5$#"(
– !"#2%"(,0&1'J%'$(5,'(&J2"#:%(5#J20%$%'%//(
• 9&P",5L#'(O,$,(5#00%5L#'N(5%'$",0(?>$(/#J%H3,$(/$,'O,"O(
– +%5#"O(J#'#53"#J,L5("%T%5L#'/(H3&0%("#$,L'1($3%(5"-/$,0(
– S>0L*H,:%0%'1$3(O,$,(5#00%5L#'(#'(/-'53"#$"#'/(
• 9,$,(2"#5%//&'1N(&'O%A&'1U(&'$%1",L#'(,'O(/5,0&'1(
• I$">5$>"%(5#'/$">5L#'N(23,/%(2"#?0%J(
– S#0%5>0,"("%20,5%J%'$V(@%,:-(,$#J(J%$3#O/((
• I$">5$>"%("%M'%J%'$(,'O(,',0-/&/N(
– +%/#0>L#'U(+*;,5$#"U(H3,$(,?#>$(?&#0#1-RW(
http://proteincrystallography.org/ & Wikipedia
456(7&,'3,'(.3%'(
E(
."-/$,00&Q,L#'(/2,5%(,'O(/5"%%'&'1N(
@#H(J>53(&/($##(J>53W(
http://www.bfsc.leidenuniv.nl/teaching
Roderick MacKinnon
Nobel Prize in Chemistry 2003
Z"#H&'1(."-/$,0(+%J,&'/([,"1%0-(,'(="$((
Y3%(/>55%//(",$%(#;(5"-/$,00&Q,L#'(
,22%,"/(?&J#O,0(R(
Try over 300 conditions is probably enough
to tell whether it will even crystallize?
http://www.ruppweb.org/Xray/tutorial/High_Throughput_EMBL_full.htm
456(7&,'3,'(.3%'(
MacKinnon, Nature (2002).
KcsA (PDB: 1r3j)
X(
456(7&,'3,'(.3%'(
D(
."-/$,0N(=/-JJ%$"&5(\'&$(,'O(\'&$(.%00((
I-JJ%$"-(
• ."-/$,0N(/#0&O/(H&$3(4$",'/0,L#',06("%2%,$/(#;(,(/-JJ%$"&5(J#L;(4>'&$(5%006(
• ='(,/-JJ%$"&5(>'&$(&/($3%(/J,00%/$(>'&$(#;(:#0>J%($3,$(5#'$,&'/(,00(#;($3%(
/$">5$>",0(&';#"J,L#'(,'O($3,$(?-(,220&5,L#'(#;($3%(/-JJ%$"-(#2%",L#'/(
5,'("%2"#O>5%($3%(>'&$(5%00](
X(5"-/$,0(I-/$%J/(
B8(2#&'$(1"#>2/(4/-JJ%$"-6(
_K(0,`5%($-2%/(4/3,2%6(
ab(8Bc(/2,5%(1"#>2/(
International symbol
(Herman-Mauguin symbol):
shortest symbols to general all
symmetry elements
e.g., P3121
P: lattice centering type (primitive)
31: symmetry of primary axis
(trigonal c-axis)
2: symmetry of secondary axis
(2 fold)
1: symmetry of tertiary axis
Restriction of chirality:
65 space groups for biomolecules!
456(7&,'3,'(.3%'(
See wikipedia for nice overview of crystal symmetry!)
^(
456(7&,'3,'(.3%'(
)*",-(9&P",5L#'(
• 9%$%"J&'%O(?-(%0%5$"#'(O%'/&$-(43-O"#1%'/(
0,"1%0-(&':&/&?0%R6(
• 9&P",5L#'N(J,A&J>J(,J20&$>O%/(H3%'(,00(
/5,d%"%O(H,:%/(,"%(&'(23,/%](
• e",11f/([,HN(4_^_E(<#?%0(!"&Q%V(g[(e",11(H,/(8E($3%']6(
Incident
I-/$%J,L5(+#$,L#'($#(!"#?%(i'L"%(+%5&2"#5,0([,`5%(
scattered
2sin "
=
#
2d sin " = n# $2sin " / # = n /d
h 2 k 2 l2
+ +
a2 b2 c 2
A single recording of diffraction, i.e., a
single reflection, at a fixed crystal
orientation only sample a limited number of
reciprocal lattice points!
Systematically rotating (a precisely aligned)
crystal allows all reciprocal lattice to be
sampled (precession imaging).
!
– ='10%(#;(O&P",5L#'(&/(h>,'LQ%O(
– +%5&2"#5,0("%0,L#'/3&2N(0,"1%"(/2,5&'1(0%,O/($#(
/J,00%"(O&P",5L#'(,'10%/(
!
_c(
Intensities of the reflections vary
dramatically, which is related to the types
and positions of atoms within the unit cell.
• S&00%"(&'O&5%/N(423)(3)$6(
– O%M'%($3%(;,J&0-(#;(0,`5%(20,'%/(
2sin "
n
h 2 k 2 l2
=
=n 2 + 2 + 2
#
dhkl
a b c
!
– :#'([,>%(ih>,L#'(
– I5,d%"&'1(:%5$#"N(42j!U((j"U($j#6(
(4"%5&2"#5,0(/2,5%R6(
456(7&,'3,'(.3%'(
Ewald Sphere (radius = 1/!)
__(
456(7&,'3,'(.3%'(
http://xray0.princeton.edu/~phil/Facility/Guides/XrayDataCollection.html
_8(
+%0,L'1(9&P",5L#'(m'$%'/&L%/(,'O(=$#J&5((
.##"O&',$%/N(I$">5$>"%(k,5$#"/(
9&P",5L#'(!,d%"'(,'O(I2,5%(Z"#>2(
• k>00-(,>$#J,L5W(
• !%,l(2&5l&'1(
• O&P",5L#'(2,d%"'(
"%5#1'&L#'(4/-/$%J,L5(
,?/%'5%/("%:%,0/(/2,5%(
1"#>26(
• ."-/$,0(>'&$(5%00(
J#"23#0#1-(
4O&J%'/&#'/6(
O%$%"J&',L#'(
• .#J20%$%'%//(
• S>0L20%(,$#J/(H&$3&'($3%(>'&$(5%00(O&P",5$(,$($3%(/,J%(,'10%/(4e",11f/(
[,H6U(?>$(H&$3(O&P%"%'$(,J20&$>O%/(,'O(23,/%/(
• I$">5$>",0(;,5$#"N(/>J(#;(/5,d%"(H,:%/(#;(,00(,$#J/((
N
F(h,k,l) = # f j e
fj: atomic scattering factor
(Xj,Yj,Zj): fractional cell coordinates
j =1
• I5,d%"%O(&'$%'/&L%/(,5$>,00-(#?/%":%ON( I(h,k,l) = F(h,k,l)
2
– m';#"J,L#'(,?#>$(23,/%(&/(0#/$R(
! • I$">5$>",0(5,05>0,L#'(;"#J()*",-(O&P",5L#'N(,'(&':%"/%(2"#?0%J(
– Y3%(;#"H,"O(2"#?0%J(#;(5,05>0,L'1($3%(O&P",5L#'(2,d%"'U(4523(3$63(;"#J(,$#J&5(
!
5##"O&',$%/(&/($"&:&,0(
– Y3%(&':%"/%(2"#5%//(#;(5,05>0,L'1(,$#J&5(/$">5$>"%U(4783983:86U(&/(<CYU(,/($3%(
5"&L5,0(23,/%(&';#"J,L#'(&/(0#/$(4&]%]U($3%(;,J#>/(23,/%(2"#?0%J6]((
• k#>"&%"($",'/;#"J(?%$H%%'("%,0(,'O("%5&2"#5,0(/2,5%/(
1
"(h,k,l) = % F(h,k,l)e #2$i(hX +kY +lZ )
": “the electron density”
V h,k,l
Indexing 1!
Cell lengths (Angstrom):
Cell angles (degree):
…!
2"i(hX j +kY j +lZ j )
6.4232
95.8000
7.3423
96.5907
15.7870!
102.5697!
amplitude (known) phase (unknown)
http://renzresearch.com/Precognition/html/chpt2_index.htm
456(7&,'3,'(.3%'(
_B(
456(7&,'3,'(.3%'(
_K(
!
.#J20%$%'%//U(=55>",5-(,'O(+%/#0>L#'((
• 9&P",5L#'/(,$(0,"1%"(,'10%/U(&]%]U(3&13%"(3U(lU(0U("%h>&"%O(;#"(3&13%"("%/#0>L#'(
4kY(>'5%"$,&'$-(2"&'5&20%6(
• S&//&'1(O&P",5L#'/(,//>J%O($#(?%(Q%"#(?-(O%;,>0$(&'($3%(/>JJ,L#'N(
"%O>5%O(,55>",5-(4,'O("%O>5%O("%/#0>L#'6(
"(h,k,l) =
1
V
% F(h,k,l)e
#2$i(hX +kY +lZ )
h,k,l
!
#$ ~ 1/T
456(7&,'3,'(.3%'(
Structural factor data F(h,k): inner
circle marks 2A resolution.
Including additional data up to the
outer circle increases resolution
to 1A (Fig 12.16 of Tinoco)
_E(
456(7&,'3,'(.3%'(
_n(
I#0:&'1($3%(!3,/%(!"#?0%J(
!3,/%jI$">5$>"%(+%M'%J%'$N(+*k,5$#"(
• =(;>'O,J%'$,0(2"#?0%J(?>$(H&$3(%/$,?0&/3%O(/#0>L#'/(
• 9&"%5$(J%$3#ON(/J,00(J#0%5>0%/(4o_G(,$#J/6(H&$3(3&13*"%/#0>L#'(O,$,(4p_(=6(
• S#0%5>0,"("%20,5%J%'$N(&;(&'&L,0(/$">5$>",0(J#O%0/(5,'(?%(%/LJ,$%O(4%]1]U(
$3"#>13(3#J#0#1-(J#O%0&'16U(!,d%"/#'(J%$3#O/(5,'(?%(>/%O($#(O%$%"J&'%O(
$3%(#"&%'$,L#'(,'O($3>/(&'&L,0(1>%//(#;(23,/%](
• m/#J#"23#>/("%20,5%J%'$N(&'$"#O>5&'1(#'%(#"(J#"%(3%,:-(,$#J/(4H&$3(0,"1%(
%0%5$"#'(O%'/&L%/6](.3,'1%/(&'(O&P",5L#'(&'$%'/&L%/(H&$3(,'O(H&$3#>$(3%,:-(
,$#J/(5,'(?%(>/%O($#(#?$,&'(&'&L,0(23,/%(%/LJ,L#'(4>/&'1(%&$3%"(O&"%5$(#"(
!,d%"/#'(J%$3#O/6](Cq%'(bB(O%"&:,L:%/('%5%//,"-](
• ='#J,0#>/(O&P",5L#'(4S=9(,'O(I=96((
http://www.sci.sdsu.edu/TFrey/Bio750/Bio750X-Ray.html
• =q%"(&'&L,0(O%$%"J&',L#'(#;(23,/%U("%/>0L'1(/$">5$>",0(J#O%0(&/(>/%O($#(
"%M'%($3%(23,/%/(,'O(5,05>0,$%(&J2"#:%O(J#O%0/(&'(,'(&$%",L:%(;,/3&#'](
• r>,0&$-(#;(M`'1(J%,/>"%O(?-(+*;,5$#"N(
Phase probability for one reflection in a MIR experiment.
(a) One derivative. (b) Three derivatives.
456(7&,'3,'(.3%'(
http://journals.iucr.org/d/issues/2003/11/00/ba5050/index.html
A 2.6 Å SIR electron-density
map with the final C% trace of
the structure superimposed.
_X(
."-/$,0(e*k,5$#"/(
– ;;"%%N(5,05>0,$%O(;"#J(p_cF(#;(O,$,('#$(
( ((((((,0"%,O-(&'50>O%O(&'("%M'%J%'$]((
– +;"%%(p("%/#0>L#'(j(_c(4&]%]U(p(c]8(;#"(8(=(/$">5$>"%6(
456(7&,'3,'(.3%'(
_D(
e*k,5$#"(,'O(.#';#"J,L#',0(k0%A&?&0&$-(
• =0/#(l'#H'(,/($%J2%",$>"%(;,5$#"(4&'50>O%O(&'(!9e(M0%/6(
• k&$($#($3%(O,$,(&'(#"O%"($#(,55#>'$(;#"($3%(s?0>""&'%//s(#;($3%(%0%5$"#'(
O%'/&$-(O>%($#($3%"J,0(J#L#'](
• e*;,5$#"/(#;(/&O%53,&'/(4E($#(nc(t86($#(?%(0,"1%"(,'O(J#"%(:,"&,?0%($3,'(
?,5l?#'%(,$#J/(4E($#(BE(t86]((
• 4\'"%0&,?0%6(&'O&5,L#'(#;(2"#$%&'(O-',J&5/W(
e*;,5$#"(a(DujB(+SIk8(
bacterial phosphotriesterase (PTE)
Correlation between loop 7 B factor and kcat in PTE variants
456(7&,'3,'(.3%'(
Jackson et al, PNAS (2009)
_^(
456(7&,'3,'(.3%'(
8c(
PDB
Molecule of the Month:
@#H(,?#>$(?&#0#1-W(
Enhanceosome
•
•
•
•
PDB: 1t2k, 2pi0,
2o6g and 2o61
Panne et al, Embo J. (2004)
."-/$,0(,"L;,5$/N(5"-/$,00&Q,L#'(5#'O&L#'/(,'O(5"-/$,0(2,5l&'1(
g3,$(,?#>$(O-',J&5/W(
I$">5$>"%(&/(v>/$(,(?%1&''&'1(w(
<S+(,'O()*",-(5"-/$,00#1",23-(&/(5#J20%J%'$,"-R(
NMR
X-ray crystallography
short time scale, protein folding
solution, purity
< 20kD, domain
functional active site
atomic nuclei, chemical bonds
resolution limit 2-3.5Å
long time scale, static structure
single crystal, purity
any size, domain, complex
active or inactive
electron density
resolution limit 1.5-3.5Å
Dynamics
Transient interactions
456(7&,'3,'(.3%'(
8_(
456(7&,'3,'(.3%'(
88(