Document 13213603
advertisement

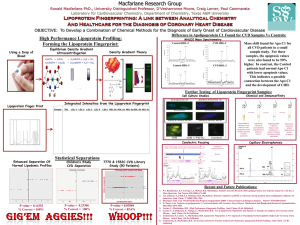
High Precision Lipoprotein Subclass Analysis (Lipoprotein TM Fingerprinting ) Ronald Macfarlane PhD., University Distinguished Professor, Craig Larner, Paul Cammarata, Catherine McNeal PhD., MD, Simon Sheather PhD. Laboratory for Cardiovascular Chemistry, Department of Chemistry, Department of Statistics, Texas A&M University, Scott & White Hospital OBJECTIVE: To test the accuracy of a new screening method for CAD on a cohort of normolipodemic subjects with & without CAD TM fingerprint How the lipoprotein of an individual is generated: Equilibrium Density Gradient Ultracentrifugation Using a Drop of Blood TM Fingerprints Utilization of Lipoprotein for Initial Clinical Studies: Objective: To determine whether lipoprotein fingerprints can be used to classify individuals with and without CAD with high accuracy. Cohorts used in clinical study: Thirty-five subjects with and without CAD were selected. Their common feature was that they all had normal lipid levels. Linear discriminate analysis ( LDA) and Sliced Inverse Regression (SIR) were selected for the classification. Statistical Separations Enhanced Separation Of Normal Lipodemic Profiles Traditional Lipid Measurement SIR/LDA Separation CVD Classifaction Long-term Objective: Develop an accurate screening method to identify individuals at high risk for coronary artery disease (CAD) prior to the development of clinical manifestations to initiate a prevention strategy. Hypotheses: •Circulating lipoprotein particles can serve as an internal probe for identifying high risk for CAD. •Effective characterization of the lipoprotein population can be improved incorporating modern methods from analytical chemistry. •Selection of lipoprotein density as a critical variable in achieving the long-term objective. •Density-gradient ultracentrifugation can be refined to obtain a high precision lipoprotein density profile compatible for clinical studies. (The profile is referred to as TM a lipoprotein fingerprint . Y N Density Gradient Theory -50 0 50 100 150 200 250 300 SIR/LDA Value H4EDTA + CdCO3 → H2CdEDTA + H2O + CO2 SIR/LDA of Traditional Lipid Measurements: P-value = 0.8852 % Correct = 50% 1.16 1.16 1.14 1.14 1.12 1.12 1.10 1.10 1.08 1.06 + 1.08 1.06 1.04 1.04 1.02 1.02 1.00 1.00 0 2 4 6 8 10 12 14 Tube Coordinate (mm) 16 18 20 0 2 4 6 8 10 12 14 16 18 20 22 Tube Coordinate (mm) Lipoprotein Fingerprints Error Reduction: Meniscus Error Removal Through Polar Layering, Enhanced Volumes Spins, and Quality Control Standards 2500 Non-CAD Sample Lipoprotein Fingerprint Prior to Enhancement: Study: 7770, Patient #142, CVD Patient 0.18M NaBiEDTA, 1150uL Spin Hexane Layered, Halide Lamp 2500 TRL 2500 TRL HDL LDL 2000 HDL LDL Mode 1 %RSD – 23.39% Mode 2 %RSD – 19.75% Intensity 2000 1500 1 2 3 4 5 2b 2c 3a 3c 3b 1 1500 2 3 4 5 2b 3a 3c 2a 3b 1000 After Enhancement: 4 1000 µL H2O Layered 1150 µL NonLayered 1150 µL Polar Layered 6 8 10 12 14 16 18 20 22 24 26 28 30 32 Tube Coordinate (mm) 0 3 6 9 12 15 18 21 24 Tube Coordinate (mm) 27 30 33 Mode 1 %RSD – 4.42% Mode 2 %RSD – 3.69% 2 3 4 5 2b 3a 3c 2a 3b 1000 0 0 500 1 1500 500 500 1000 Intensity 2000 Intensity TRL Non-CAD Contributor – Green n Importance of Subclasses References: -0.15 -0.10 -0.05 0.00 0.05 0.10 0.15 0.20 SIR/LDA Values SIR/LDA of Mode 2: P-value = 0.03290 % Correct = 100% Conclusion: HDL LDL y -0.20 CVD Sample Lipoprotein Fingerprint Study: 1532C, Patient 001, Angiogram Based Control 0.18M NaBiEDTA, 1150uL Spin Hexane Layered, Halide Lamp 10 HPLPD Profiles of a Serum Sample - Overlaid 1150uL 0.18M NaBiEDTA, 6uL Serum, 10uL NBD C6-Ceramide, 200+ uL Hexane Layered Weighted Constants for SIR/LDA Prediction Lipid Profiling Measurements - Mode 2 SIR/LDA Separation CVD Classification Density (g/mL) Density (g/mL) H2CdEDTA + Cs2CO3 → Cs2CdEDTA + H2O + CO2 5 10 15 20 25 30 Tube Coordinate (mm) CAD Contributor - Red SIR/LDA Value = 2.218 + 0.014 x Ln(TRL) + 0.042 x Ln(LDL-1) + -0.124 x Ln(LDL-2) + -0.129 x Ln(LDL-3) + -0.088 x Ln(LDL-4) + 0.336 x Ln(LDL-5) + -0.645 x Ln(HDL-2b) + 0.279 x Ln(HDL-2a) + -0.448 x Ln(HDL-3a) + 0.280 x Ln(HDL-3b) + -0.269 x Ln(HDL-3c) •HDL subclasses play a significant role in classifying CAD/ no CAD individuals with normal lipid levels with 100 % accuracy. •TG, HDL-c, LDL-c levels alone cannot classify CAD/no CAD individuals with normal lipid levels. •The high precision of the Lipoprotein Profiles is CRITICAL to classification of samples and how it works. •Macfarlane, R. D.; Hosken, B. D.; Farwig, Z. N.; Espinosa, I. L.; Myers, C. L.; Cockrill, S. L. Lipoprotein fingerprinting method. U.S. Patent 6,753,185, 2004 •Hosken, B. D.; Cockrill, S. L.; Macfarlane, R. D. Anal. Chem. 2005, 77, 200-207 •Johnson, J.D.; Bell, N.J.; Donahoe, E.L.; Macfarlane, R.D. Anal. Chem. 2006, 77, 7054-7061 •Espinosa, I. L.; McNeal, C. J. ; Macfarlane, R. D. Anal. Chem. 2006, 78 (2): 438-444 •Henriquez, R.; Chandra, R.; Hosken, B. D.; Macfarlane, R. D. Atheroscler. Suppl. 2006, 7 (3): 587-588 •Larner, C.L.; Henriquez, R.H.; Macfarlane, R.D.; McNeal, C.J.; Sheather, S. 15th International Symposium on Atherosclerosis, Boston, MA, June 14-18, 2009; International Atherosclerosis Society: Milan, Italy, 2009; P790. This project was funded by NIH/NHLBI, grant number RO1 HL068794, and Scott & White Hospital.