Understanding bacterial regulation systems to unlock
advertisement

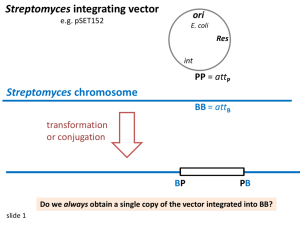
Understanding bacterial regulation systems to unlock the production of novel “cryptic” antibiotics Vincent Poon1,2, Dr. Jonathan Moore1 and Dr. Christophe Corre2 Vincent.Poon@Warwick.ac.uk 1Warwick Systems Biology Centre, University of Warwick 2School of Life Sciences / Department of Chemistry, University of Warwick Aims of the project • To understand regulatory systems involved in controlling production of antibiotic-like compounds in Streptomyces bacteria. • To exploit our understanding to unlock the production of novel antibiotics. 1. Introduction Up to two thirds of today’s clinically approved antibiotics originate from soil-living Streptomyces bacteria (Fig 1) [1]. While each species is predicted to produce dozens of drug-like compounds, the production of compounds is often tightly controlled by DNA-binding regulators (Fig 2). Fig 1. Streptomyces secreting blue pigmented antibiotic 2. Antibiotic-producing gene clusters controlled by conserved regulatory systems In diverse Streptomyces bacteria, the production of drug-like compounds is regulated by a conserved set of 5 genes. These regulatory genes are clustered with genes that direct the assembly of antibiotic-like compounds (Fig 2) [2]. By understanding how the production of compounds is regulated, genetic manipulations can be carried out in these gene clusters to discover new natural drug O compounds. Streptomyces coelicolor 12kb O HO2C Methylenomycin (19,342 bp) Streptomyces venezuelae 3kb O (11,670 bp) Gaburedin Streptomyces avermitilis 35kb 1 kb (39,616 bp) Key: DNA-binding Repressor Target sequence of the repressors Signal assembly CO2H N H N H CO2H Activator Fig 2. Gene clusters involved in regulating the production of drug-like compounds in Streptomyces bacteria 3. Predicted novel valanimycin-like compounds in Streptomyces avermitilis gene cluster 1 kb Streptomyces avermitilis gene cluster Streptomyces viridifaciens (valanimycin producer) Valanimycin ? Fig 3. Streptomyces avermitilis gene cluster is predicted to produce a valanimycin-like compound. Interestingly, all of the essential genes involved in the production of the valanimycin natural product (antibacterial and anticancer compound), from Streptomyces viridifaciens, are also present in Streptomyces avermitilis (Fig 3 – N+ N -O CO2H identical genes are colour coded). Streptomyces avermitilis is proposed to produce a valanimycin-like compound. 4.Synthetic biology - “Playing” with regulators 5. Conclusions and Future works In Streptomyces coelicolor and Streptomyces venezuelae (Fig 2), the An untapped source of drug compounds is waiting to be deletion of regulators over-produces the compound. The rational genetic discovered in Streptomyces [2]. Using a synthetic biology manipulation of the homologous regulator in Streptomyces avermitilis is approach, Streptomyces avermitilis is being manipulated expected to over-produce new valanimycin-like compounds. and is expected to produce valanimycin-like compounds. References [1] Bentley, S.D. et al. (2002). Nature. [2] Corre, C. et al. (2008). Proc. Natl. Acad. Sci. [3] Ling, L.L. et al. (2015). Nature. Acknowledgements The authors would like to acknowledge BBSRC and Systems Biology DTC for funding and the Royal Society of Chemistry for funding the laboratory. Visit my ePortfolio