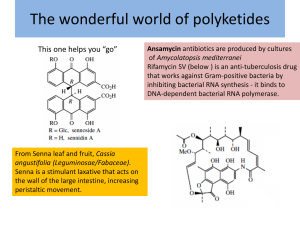
David Hopwood (Lecture 1)
advertisement

David Hopwood Lecture 1 (DH1) Isolation of microbes from soil: fungi, actinomycetes, other bacteria (left); streptomycetes (right) Bioactive compounds from microbes (2002) Antibiotics Other Total Actinomycetes 7900* 1220 9120 Other bacteria 1400 240 1640 Fungi 2600 1540 4140 Total 11,900 3000 14,900 *70% from Streptomyces spp. Actinomycetes Other bacteria Fungi Diminishing returns in finding useful natural products Antibiotic producers are differentiating microbes Penicillium notatum (penicillin) Aspergillus terreus (lovastatin) Penicillins Cephalosporins Griseofulvin Valuable fungal metabolites Cyclosporin Lovastatin Some myxobacteria Myxococcus Stigmatella Sorangium (epothilone) Myxobacterial metabolites Epothilone Et Me Me R O OH R R R E S E S E Valuable myxobacterial metabolites Me S Me O R OH S S CO 2 H Ambruticin A Streptomyces colony on an agar plate Streptomyces: scanning EM Young vegetative hyphae Transition stage: most antibiotic production Aerial hyphae, young spores Mature spores Reproduction Antibiotic production Apoptosis: nutrient release Feeding Fatal attraction Needs for new antibiotics •Overcome acquired resistance: Staphylococcus aureus (MRSA) Vancomycin-resistant Enterococcus MDR and XDR Mycobacterium tuberculosis Gram-negative respiratory pathogens •Less toxic anti-viral or anti-cancer agents •Immunosuppressants, cholesterol lowerers… How to find new antibiotics •Novel natural products •Chemical synthesis combichem •Genetics, genetic engineering Genetics of antibiotic producers Filamentous fungi: gene replacements, genomics Myxobacteria: transduction, transposon libraries, gene replacements, genomics… Streptomyces: plasmid-mediated conjugation, protoplast fusion, autonomous and integrating plasmid and phage cloning vectors, gene replacements, transposon libraries, genomics… Streptomyces cloning vectors Class Replicon Example Low copy number autonomous plasmid SCP2* pRM5 High copy number autonomous plasmid pIJ101 pIJ699 Single copy number integrating plasmid pSAM2 None pPM927 pSET152 Single copy number integrating phage ΦC31 KC515 Streptomyces plasmid SCP2 Streptomyces phage C31 Streptomyces mycelium and protoplasts, light microscope Streptomyces mycelium and protoplasts, electron microscope The Streptomyces coelicolor genome 1958 First Streptomyces coelicolor linkage map First antibiotic gene (later named act) 1965 antibiotic bald white 19901990 1993 (325 clones) Http://jic-bioinfo.bbsrc.ac.uk/streptomyces - then click “ScoDB II” LH arm = 1.5 Mb RH arm = 2.3 Mb 9 May 2002 Core = 4.9 Mb 7825 ORFs (55 pseudogenes) 63 tRNA genes 6 rRNA operons 72.12% G+C Isolation of antibiotic biosynthetic genes act mutant of Streptomyces coelicolor (Brian Rudd, 1976) act mutants of Streptomyces coelicolor The first act clone (Francisco Malpartida, 1984) The act genes of Streptomyces coelicolor Tailoring steps Regulation resistance Actinorhodin Chain assembly (PKS) Medermycin (S. AM-7161) Actinorhodin (S. coelicolor) Mederhodin First ‘hybrid’ antibiotic (1985) Manipulation of polyketide biosynthesis Some actinomycete antibiotics (Polyketides) Medicine Application Examples Agriculture Application Examples Anti-bacterial Erythromycin Livestock Tetracyclines rearing Rifamycin Anti-cancer Adriamycin Monensin Tylosin Virginiamycin Anti-parasitic Avermectin ImmunoFK 506 suppression Fungicide Polyoxin Kasugamycin Antifungal Herbicide Bialaphos Candicidin 6-MSA Cyanidin Polyketides Aflatoxin Oxytetracycline Erythromycin Brevetoxin A fatty acid Palmitic acid COOH Variables in polyketides (‘Combinatorial biosynthesis’) Extender Chirality Side chains O Starter R OH O OH OH Reduction level KR/DH/ER H O Chain length The act and ery PKS gene clusters KS Type II PKS act (simple) CLF * * Type I modular PKS ery (complex) DEBS1 LM Module 1 AT KR AT ACP KS Module 2 Module 3 S DEBS3 Module 5 Module 4 DH ER AT KR ACP KS S O DEBS 2 ACP KS AT ACP KS AT S S AT KR KR ACP KS Module 6 TE AT KR ACP KS S O O OH ACP TE S O S O O O O O HO HO O HO HO HO HO O HO HO O HO HO O HO HO 1 O OH OH 6dEB HO HO The DEBS paradigm for complex polyketide biosynthesis Discovery of ‘cryptic’ secondary metabolites ‘Secondary metabolic’ gene clusters in Streptomyces coelicolor 3 antibiotics (type II PK, modular PK, NRP) 4 siderophores (2 NRP, 2 other) 3 pigments (type II PK, chalcone, carotenoid) 2 complex lipids (unsaturated FA, hopanoid) 2 signalling molecules (terpenoid, -butyrolactone) 8 other (2 modular PK, 1 NRP, 2 chalcones, 2 terpenoid, 1 deoxysugar) PK = polyketide, NRP = non-ribosomal peptide, FA = fatty acid Total length ~ 375 kb ~ 4.5% of the genome S. coelicolor v. S. avermitilis Class S. coelicolor S. avermitilis Type I PK 3 8 Type II PK 2 (1) 3 (1) NRP 4 6 Carotenoid 1 1 Desferrioxamine 1 1 Chalcone 3 (1) 1 Others 9 4 Red = similar gene clusters Enediynes PKS Zazopoulos et al. (2003) Nature Biotech. 21:187 Discoveries/year Cumulative discoveries Watve et al. (2001) Arch. Microbiol. 176:386 “How many antibiotics are produced by the genus Streptomyces?” 500 current effort level 2003 Total may be 150,000! “Therefore, by genic manipulation of the cell we have a means for obtaining, in quantities sufficient for study, many of the metabolic products of the living organism that would be otherwise undetectable” Albert Kelner (1949) Improvement of productivity Competing pathways Cofactor availability Shunt products Pathway genes Uptake Export Product Substrate Synthesis Undesired substrate Feedback inhibition + Regulation Need for functional genomics Environmental factors Some targets for influencing antibiotic productivity Many genes with cumulative effects! 22,000 survivors of mutagenesis Pick 11 best strains and fuse protoplasts Pick 7 best from 1000 progeny and fuse protoplasts Screen 1000 progeny: 2 as good as the best from 1 million cultures screened over 20 year [Zhang, y. et al. (2002) Nature 415: 644] Increased productivity of S. fradiae for tylosin (24,000 colonies screened over 1 year)