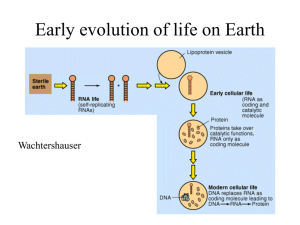
Document 12023055
advertisement

AN ABSTRACT OF THE THESIS OF Michael A. Haddad for the degree of Master of Science in Microbiology presented on August 18, 1994. Title: Phylogenetic Characterization of the Epibiotic Bacteria Associated with the Hydrothermal Vent Polychaete Alvinella pornpejana. Abstract approved: Redacted for Privacy Stephen J. Giovannoni Symbiotic relationships of bacteria with higher organisms are commonly observed in nature; however, the functional role of these relationships is only rarely understood. This is particularly evident in epibiotic bacterial associations in the marine environment where the bacteria are often a diverse ensemble of microorganisms, thus complicating the identification of the functionally important members. Classical microbiological techniques, relying primarily on culturing these organisms, have provided an incomplete picture of these relationships. Molecular genetic techniques, focusing on the analyses of bacterial 16S rRNA sequences cloned directly from natural microbial populations, are now available which allow a more thorough examination of these associated bacterial populations. This study sought to characterize the epibiotic bacterial population associated with a very unique organism, Alvinella pompejana, using such a molecular approach. Alvinella pompejana is a polychaetous annelid that inhabits active deep-sea hydrothermal vent sites along the East Pacific Rise. This worm colonizes the walls of actively venting high temperature chimneys and is thought to be one of the most thermotolerant metazoans known. The chimney environment is characterized by high concentrations of sulfide and heavy metals in the vicinity of the worm colonies. A morphologically diverse epibiotic microflora is associated with the worm's dorsal integument, with a highly integrated filamentous morphotype clearly dominating the microbial biomass. It has been suggested that this bacterial population participates in either the nutrition of the worm or in detoxification of the worm's immediate environment; however, previous studies have been unable to confirm such a role. The primary goal of this study is to phylogenetically characterize the dominant epibionts through the analysis of 16S rRNA gene sequences. Nucleic acids were extracted from bacteria collected from the dorsal surface of Alvinella pompejana. 16S rRNA genes were amplified with universal bacterial primers by the polymerase chain reaction (PCR). These genes were subsequently cloned and the resulting clone library was screened by restriction fragment length polymorphism (RFLP) analysis to identify unique clone types. Thirty-two distinct clone families were found in the library. Four of these families were clearly dominant, representing over 65% of the library. The main assumption in this study is that the numerical dominance of the phylotypes in the starting population will be reflected in the clone library. Thus, representative clones from the four most abundant clone families were chosen for complete gene sequencing and phylogenetic analysis. These gene sequences were analyzed using a variety of phylogenetic inference methods and were found to be related to the newly established epsilon subdivision of the Proteobacteria. In future studies, these gene sequences will be used to construct specific oligodeoxynucleotide probes which can be used to confirm the morphology of the clone types in the epibiont population. Phylogenetic Characterization of the Epibiotic Bacteria Associated with the Hydrothermal Vent Polychaete Alvinella pompejana by Michael A. Haddad A THESIS submitted to Oregon State University in partial fulfillment of the requirements for the degree of Master of Science Completed August 18, 1994 Commencement June 1995 Master of Science thesis of Michael A. Haddad presented on August 18, 1994 APPROVED: Redacted for Privacy Major Prof ssor, re pr enting Microbiology Redacted for Privacy Chair Department icrobiology Redacted for Privacy Dean of Graduate School I understand that my thesis will become part of the permanent collection of Oregon State University libraries. My signature below authorizes release of my thesis to any reader upon request. Redacted for Privacy Michael A. Haddad, Author ACKNOWLEDGEMENTS I would like to first acknowledge the Molecular and Cellular Biology program here at Oregon State University for providing the financial support for my first year of study. I also would like to thank everyone in my lab for their encouragement and support. I am especially grateful to both Craig Cary and Steve Giovannoni for giving me the opportunity to work on such an exciting project and the guidance to help me see it through to completion. There are so many more people whom I could acknowledge here, the most important of which is my family. I have been extremely lucky to have such a supportive and generous group of people as my main support system. First my mother, Niki Ross, has been fundamental to the development of myself as a human being. If there is one person whom I have learned the most from about life it is her. My step-father, Bill Ross, has always encouraged me about everything I have ever done (except maybe the mohawk). My father, Sam Haddad, has perhaps given the gift I will treasure the most, an education. He has financially supported my education with out ever setting any limits. As a result, he has allowed me to pursue whatever I have wanted. That is probably one of the most important freedoms to ever have. Finally, I am thankful for the tremendous amount of support I have had from my friends: Bill Idel, Greta Wade, Becky Bierenbaum, and Jacob Benjamin Perkinson. Without question, the most important person I have to thank is my partner, Laura MacGregor. She is the one who has dealt with all of my trials and tribulations first-hand, and has always been there for me. For that enormous amount of support I am eternally grateful. TABLE OF CONTENTS INTRODUCTION 1 Ecology of Alvinella pompejana The Epibiotic Microflora of Alvinella pompejana Possible Role of the Epibionts 3 Conclusions of Previous Research 13 Use of the 16S rRNA Molecule Experimental Rationale 14 15 METHODS Sample Collection 6 11 18 18 Bacterial Removal and DNA Purification. 18 Amplification of 16S rRNA Genes 19 Construction and Screening of Clone Library. RFLP Analysis. Gene Sequencing Phylogenetic Analysis 20 RESULTS Clone Library Construction Restriction Fragment Length Polymorphism Analysis 16S rRNA Gene Sequences Phylogenetic Analysis 21 22 23 25 25 25 26 29 DISCUSSION 39 BIBLIOGRAPHY 45 APPENDICES 52 LIST OF FIGURES Figure 1. 2. 3. 4. 5. 6. 7. Page Map showing active hydrothermal vent sites along the East Pacific Rise (EPR) 5 Dorsal view of the holotype of (a) Alvinella pompejana and (b) Alvinella caudata 8 Schematic outline of the experimental strategy used for the phylogenetic analyses of the epibiotic microbial community associated with the hydrothermal vent polychaete Alvinella pompejana 17 Scaled Illustration of the restriction patterns of 16S rDNAs representing 11 clone families identified in the alvinellid clone library. 27 Pylogenetic tree showing the relationship of the alvinellid clones to the other members of the Proteobacteria. 30 Proposed secondary structural model of clone APG 13B 16S rDNA 33, 34 Proposed secondary structural model of clone APG 56B 16S rDNA. 35, 36 LIST OF TABLES Table 1. 2. 3. Page Chemical composition of vent end-member fluid @ 21° N on the East Pacific Rise vs. ambient sea water 7 RFLP analysis results showing the numberical distribution of the alvinellid clone families. 28 Signature base positions distinguishing the delta and epsilon subclasses of Pro teobacteria and the alvinellid clones. 38 LIST OF APPENDICES Page Appendix 1: Locus APG 5A 1496 by DNA 52 Appendix 2: Locus APG 13b 1502 by rRNA 54 Appendix 3: Locus APG 44b 1502 by DNA 56 Appendix 4: Locus APG 56B 1504 by DNA 58 Phylogenetic Characterization of the Epibiotic Bacteria Associated with the Hydrothermal Vent Polychaete Alvinella pornpejana INTRODUCTION Integrated bacterial associations with animals are widely distributed in both terrestrial and marine systems. Although many of these relationships appear obligatory, very little is known about the functional role of the bacteria. As a result, whether these bacteria reside intracellularly (endosymbiotic) or extracellularly (episymbiotic or epibiotic), many of these affiliations can only roughly be defined as symbiotic. It has been impossible to determine whether they are mutualistic, where both members are benefited by the relationship, or merely commensal, where only a single member profits. Epibiotic associations of bacteria with metazoans are common in the marine environment. Many of the affiliations are monospecific and invariant in nature, often involving a single bacterial species with a specific host group. Bioluminescent bacteria colonizing the light organ of the sepiolid squid Euprymna scolopes comprise a single strain of Vibrio fischeri (McFall-Ngai et al., 1991) However, more frequently these epibiotic assemblages are composed of a phenotypically diverse group of microorganisms, which further complicates resolving the contribution of each individual member with its host. Several distinct morphological types of bacteria are found colonizing the gills of the rock crab Cancer irroratus and are believed to play an important role in the pollution of these organisms (Bodammer et al., 1981). Similarly, periphytic bacteria are associated with another crab, He lice crassa, found at tannery effluent sites, and may contribute 2 to the concentration of chromium in these environments (Johnson et al., 1981). An unnamed limpet species inhabiting deep-sea hydrothermal vent sites is also found with dense colonies of filamentous bacteria colonizing the gill epithelial surface (de Burgh et al., 1984). Probably one of the more dramatic examples of invertebrate/bacteria associations are those found in certain sponges. In the majority of the large marine sponges, dense populations of bacteria reside in the intercellular spaces, comprising up to one-third of the volume of the sponge. These bacterial populations are typically composed of between 4-7 different morphological types (Smith et al, 1987). Although there have been several attempts to isolate these bacteria free from their hosts, few have been confirmed to be the symbionts. In these and other similar associations the task of distinguishing and characterizing the important members in these epibiotic associations from those opportunistic bacteria, has remained, until recently, intractable. One of the more remarkable members of deep-sea hydrothermal vent communities is the polychaetous annelid, Alvinella pompejana, found colonizing the walls of actively venting chimneys (Desbruyeres et al., 1980). A highly diverse and dense assemblage of microorganisms, including an integrated filamentous morphotype, occupies the dorsal surface of this worm (Desbruyeres et al., 1985; Gaill et al., 1987). To date, this filamentous morphotype has eluded all attempts at culturing (Durand et al., 1990; Jeanthon et al., 1992; Prieur, et al. 1990). Several studies have suggested that the bacteria participate in the nutrition of the host or in the detoxification of the host's immediate environment (Alayse-Danet et al., 1987; Gaill et al., 1988). However, due to the inherent difficulty in working with A. pornpejazza, which rarely survives trips to the surface, the apparent failure to 3 culture the dominant morphotypes, these hypotheses have remained merely suggestive. Molecular genetic strategies now exist for the characterization of microorganisms which have previously not been observed in culture. The analysis of small subunit ribosomal RNA sequences (16S rRNA) cloned directly from natural populations has expanded the limits of microbial ecology and diversity over that which was possible with more traditional microbiological techniques. In other studies, 16S rRNA sequence analysis has provided a powerful means to examine the diversity of natural microbial populations in marine and fresh water systems avoiding the reliance on cultavatibility. These same techniques can be used to phylogenetically characterize the microflora associated with Alvinella pompejana. Characterizing the dominant members of the microflora would provide essential information necessary to determine the role of these bacteria. These same techniques can also be applied to other epibiotic microbial associations, which will no doubt add to our limited knowledge about these other associations. Ecology of Alvinella pompejana Alvinella pompejana is a tube-dwelling, polychaetous annelid that colonizes actively venting chimneys at deep-sea hydrothermal vent sites (Desbruyeres & Laubier 1980). A. pompejana, as well as its congener A. caudata, are placed within their own family, the Alvinellidae (Desbruyeres et al., 1986), along with another genus Paralvinella (Desbruyeres et al., 1985). All members of the family appear to be restricted solely to deep-sea hydrothermal vent sites (Desbruyeres et al., 1991). The Alvinellidae range in distribution 4 from ITS to 50°N along the East Pacific Rise (EPR) in the eastern Pacific Ocean (Figure 1), with Alvinella spp. being restricted to the high temperature vent sites from 17°S-21°N (Desbruyeres & Laubier, 1991; Tunnicliffe et al., 1993). Alvinella spp. form dense colonies along the walls of actively venting chimney-like mineral structures. These worms build very stable tubes composed of both inorganic and organic material which are cemented to the chimney walls (Gaill et al., 1986; Vovelle et al., 1986). It has been suggested that these tubes assist in the stabilization of venting chimney structures by providing additional structural substrate for the further deposition of mineral precipitates (Tunnicliffe et al., 1990). The immediate environment of Alvinella pompejana is one of the most extreme environments on the planet. A. pompejana is one of the few invertebrate species present at vent sites to live in close proximity to the hot hydrothermal vent effluent. These worms are found colonizing basal mounds, zinc-sulfide and black smoker chimneys where the core effluent temperature ranges from 80°C-350°C, depending on the chimney type (Desbruyeres & Laubier, 1991). The mixing zone of the hot hydrothermal fluid with the cold ambient seawater produces a sharp temperature gradient, ranging from 350°C - 1.8°C within a few centimeters (Desbruyeres et al., 1985). In one study, temperature readings within an alvinellid colony were 20-35°C at the tube opening, 100°C 10 cm inside the tube mass, and 249°C 20 cm inside the tube mass (Desbruyeres et al., 1982). Recently, an actively swimming specimen of A. pompejana was observed resting on the submersible's temperature probe, which simultaneously registered a temperature of 105°C. This observation lead the authors to suggest that A. pompejana may hold the record for the upper 12' 49 N 12' 48 Figure 1. Map showing active hydrothermal vent sites along the East Pacific Rise (EPR). (a) The Alvinellidae range in distribution from 17°S to 50'N along the EPR, with Alvinella spp. being restricted to the high temperature vent sites from 17°S-21°N. (b) Alvinella pompejana specimens used in this study were collected from the Elsa vent field at the 13°N site. This figure was adapted from: Jollivet, D. 1993. Distribution et evolution de la faune associee aux sources hydrothermales profondes a 13°N sur la dorsale du pacifique oriental: le cas particulier des polychetes alvinellidae. These de doctorat de L'Universite de Bretagne Occidentale. IFREMER, Centre de Brest. 6 temperature limit of any eukaryote (Chevaldonne et al., 1992). Although additional data are required to confirm this claim, it is believed that these organisms exist at environmental temperatures between 20°C and 60°C (Desbruyeres & Laubier, 1991). Another consequence of colonizing active chimneys at deep-sea hydrothermal vent sites is the direct exposure to high concentrations of heavy metal compounds and hydrogen sulfide contained in the vent effluent. Elevated concentrations of these compounds are well known to inhibit the growth and essential biochemical activities of microorganisms and eukaryotes (Collins et al., 1989; N.R.C., 1979). Table 1 shows concentrations of some of these compounds at a site at 21'N on the East Pacific Rise in comparison to average seawater. As is evident from this table, the vent water is acidic and enriched in hydrogen sulfide and biologically toxic heavy metals, such as arsenic, lead, and silver. Due to the proximity of the worm's tubes to the vent effluent, A. pompejana and its associated microflora are constantly exposed to these toxic compounds (Desbruyeres et al., 1985; Desbruyeres & Laubier, 1991). The Epibiotic Microflora of Alvinella pompejana A fascinating feature of both A. pompejana and A. caudata is the presence of an epibiotic population of bacteria associated with the worms' dorsal surface (Figure 2). These alvinellid species differ somewhat in the structural features that link the epibionts to their dorsal surface. A. pompejana posses modified dorsal digiform expansions heavily colonized by filamentous bacteria. Similar filamentous bacterial morphotypes are linked to modified posterior parapodia in A. caudata (Desbruyeres & Laubier, 1991). 7 Table la. Chemical composition of vent end-member fluid at 21°N on the East Pacific Rise vs. ambient sea water. 21 °N Ambient sea water Li (gM) 891 Na (mM) K (mM) 432 23.2 26 464 9.79 Rb (p.M) 28 1.3 Be (nM) Mg (mM) 15 0.02 0 52.7 Ca (I_LM) 15.6 10.2 Sr (g.M) 81 Ba (11M) pH >7 3.4 87 0.14 Cl (mM) Si (mM) 489 17.6 0.16 Al (11M) 5.2 0.01 SO4 (mM) H2S (mM) 0 27.9 0 NH3 (mm) Mn (gM) 7.3 0 960 <0.001 Fe (i.i.M) 1664 <0.001 Co (nM) 213 0.03 Cu (p.M) 35 106 0.007 38 0.02 155 1.0 308 247 0.01 Zn (tiM) Ag (nM) Cd (nM) Pb (nM) As (nM) Se (nM) 72 7.8 541 0 0.01 27 2.5 a Table was taken from Edmond, J. M. and K. L. Von Damm. 1985. Chemistry of ridge crest hot springs. In: M. L. Jones (Ed.), The hydrothermal vents of the eastern Pacific: an overview. Bull. Biol. Sci. Wash. 6:43-47. Figure 2. Dorsal view of the holotype of (a)Alvinella pompejana and (b) Alvinella caudata. Arrows indicate approximate positionS of bacterial sampling for this study. This figure was adapted from: Desbruyeres, D. and L. Laubier. 1980. Alvinella pompejana gen. sp. nov., ampharetidae aberrant des sources hydrothermales de la ride Est-Pacifique. Oceanol. Acta 3: 267-274. co 9 In both species the bacteria appear as a gray mat covering the majority of the worms' surfaces and are also found lining the interior of the worms' tube (Desbruyeres et al., 1985; Gaill & Hunt, 1986). The density of the bacterial population seems to increase posteriorly, presumably responding to some environmental gradient (Desbruyeres et al., 1980; Gaill et al., 1987). For the purposes of this discussion, only the details of the epibiotic microbial population associated with A. pompejana will be further emphasized. The epibiotic microbial population associated with Alvinella pompejana is morphologically diverse. Electron microscopic observations have documented many morphotypes, including rod, coccoid, spiral, and filamentous bacteria (Gaill et al., 1987). Although it is impossible to accurately identify bacterial species based solely on morphology, these studies have served to document several interesting patterns of the distribution of these morphotypes in the population. These morphological forms have been found in several different distributions on the host, ranging from rod-shaped bacteria found randomly dispersed, to filamentous bacteria found colonizing specific locations on the worm (Desbruyeres et al., 1985; Gaill et al., 1987). These filamentous morphotypes dominate the population representing over 60% of the microbial biomass (Gaill et al., 1987). Filamentous bacteria are found joined to small expansions of the worm's epidermis within the intersegmentary spaces (Gaill et al., 1987). These expansions are 10 tm in length and appear composed of glandular cells (Desbruyeres et al., 1985). The filamentous bacteria are embedded in a polysaccharide secretion from the worm (Gaill et al., 1988), and have been shown to be in an active state of growth (Alayse-Danet et al., 1986). The appearance of this specific association and abundance of the morphological form in the population has lead others 10 to suggest a specific relationship between these filamentous bacteria and A. pompejana (Gain et al., 1988). The high diversity of the epibiotic microbial community associated with Alvinella pompejana is apparent not only from the morphology, but also in the wide range of metabolic capabilities detected in the community. Several attempts have been made at culturing the epibiotic bacteria free from A. pompejana. A preliminary study of the heterotrophic bacteria present in this community yielded 62 total isolates. All of the isolates were Gram- negative and rod-shaped in their morphology (Prieur et al., 1987). In later studies additional heterotrophs and several autotrophic bacteria were successfully isolated. The majority of these new isolates had resistance to one or more selected heavy metals, such as silver, copper, cadmium, zinc, and arsenic (Jeanthon et al., 1990; Prieur et al., 1990). Numerous sulfur-oxidizing and sulfur-reducing isolates were also obtained, as well as several isolates involved in the nitrogen cycle, and in the oxidation of manganese (Durand et al., 1990). Growth of these isolates was observed at temperatures ranging from 20°C to 80°C, at both atmospheric and in situ pressure (250 atm). Collectively, these studies complement early microscopy findings by demonstrating the microflora of A. pompejana is both morphologically and metabolically diverse. In all of these studies the dominant filamentous morphotype was never confirmed to be in culture, thus illustrating the limitations of these classical approaches. 11 Possible Role of the Epibionts Several studies have speculated on the role of the epibiotic microbial community in the survival of A. pompejana. The primary hypothesis has been that the microflora in some way participates in the nutrition of the alvinellid, either by the worm directly feeding on the bacteria, or by a translocation of specific metabolites from the bacteria to the worm (Gaill et al., 1988). A. pompejana has a complete digestive system similar to other ampharetids (Saulinier-Michel et al., 1990). Analysis of both the mouth structures and lumenal contents reveal the presence of bacterial cells (Desbruyeres et al., 1983). This suggests that direct feeding on bacteria does occur, but its significance to the overall nutrition of the host remains unknown. Comparison of 513C values of A. pompejana and Rifitia pachyptila, a gutless, vent invertebrate that harbors endosymbiotic chemoautotrophic bacteria, suggests that these two organisms derive their organic carbon from a similar source (Desbruyeres et al., 1983). The nutrition of Riftia pachyptila is provided entirely by the resident chemoautotrophic endosymbionts (Cavanaugh et al., 1981; Felbeck, 1981). At this time it is unknown whether the endosymbionts provide nutriment via some translocation product or are merely selectively digested by the host. Examinations of the worm's anatomy reveals disorganized collagenous fibers underlying the worm's cuticle at the intersegmentary spaces (Gaill et al., 1987) as well as highly vascularized underlying epithelium (Desbruyeres et al., 1985). Some of the rod-shaped bacteria present in the population have thin filamentous structures which attach to the worm's cuticle (Gaill et al., 1987). These observations have led 12 the authors to suggest the potential for the passage of metabolites from the bacteria to the worm (Alayse-Danet et al., 1987; Gain et al., 1988). A second hypothesis for the role of the epibionts is in the detoxification of the immediate environment of Alvinella pompejana (Alayse-Danetet al., 1987; Gain et al., 1988). As stated previously, this alvinellid lives in an environment with high concentrations of biologically toxic heavy metals and hydrogen sulfide. Many bacteria are known to possess strategies for toxic heavy metal resistance, such as sequestering metals in the cell wall or in intracelluar compartments, enzymatic modification of these metals to less toxic forms, or by rapidly expelling these metals from the cytoplasm (Silver et al., 1989). It is thought the epibiotic microflora of A. pompejana could participate in the detoxification the worms' environment through the concentration of toxic heavy metals. A similar function for the epibiotic bacteria of the crab He lice crassa has also been proposed (Johnson et al., 1981). These bacteria colonize the carapace and gills of the crab and participate in the concentration of chromium. Hydrogen sulfide, at concentrations less that one-thousandth that found in vent habitats, is highly toxic to almost all living systems. Sulfide poisons certain respiratory proteins, such as cytochrome c oxidase (N.R.C., 1979), involved in the use of molecular oxygen in aerobic respiration. Several novel mechanisms exist in certain invertebrates to cope with the elevated sulfide levels in the vent systems. Riftia pachyptila possesses a sulfide binding protein in the blood which tightly binds free sulfide thus preventing access to sulfide sensitive cellular constituents (Arp et al., 1983; Powell et al., 1983). Marine organisms that are routinely exposed to toxic levels of locally produced sulfide often possess high activities of sulfide oxidizing systems (Somero et al., 1983). Although the blood of A. pompejana possesses 13 extremely high binding efficiencies for oxygen in order to deal with the elevated temperatures and hypoxic conditions (Toulmond et al., 1990), no specific sulfide binding capability has been reported. It is conceivable that the microbial community associated with A. pompejana oxidizes available sulfide within the tube, thereby detoxifying the immediate environment of the worm. Conclusions of Previous Research The majority of epibiotic associations of bacteria with metazoans are poorly understood. Deciphering the functional role of the bacteria in these relationships is often confounded by the complexity of the associated bacterial communities. Nowhere is this problem more evident than with the epibiotic bacterial community associated with the deep-sea hydrothermal vent polychaete A. pompejana. This epibiotic microflora has been shown to be both morphologically and metabolically diverse; however, the exact relationships between the bacteria and the polychaete have yet to be definitively established. It has been suggested that the bacteria participate in the nutrition of the worm, and/or in the detoxification of the worms' environment. Previous experiments have been unable to prove either of these hypotheses. One of the main difficulties in researching such a relationship is the viability of the microorganisms in culture. The filamentous bacterial form, which has been suggested to play an important symbiotic role, has not been observed in previous culture attempts. It is conceivable that these bacteria are pleiomorphic and might already be in previously established culture collections, however the reliance on morphology for the characterization of this population is a major weakness 14 in these studies. Even if the pleiomorph of the filamentous bacteria did appear in culture, it would be difficult to identify its spatial distribution in the worm's environment. In order to circumvent these difficulties and begin to understand the phylogenetic diversity of the alvinellid epibiont community, an experimental strategy developed for the analysis of mixed populations of free-living microorganisms was chosen. This approach is based the philosophy that, like organelles, microorganisms can be studied without cultivation. The approached used in this study is based on the sequence and phylogenetic analysis of bacterial 16S rRNA genes cloned directly from natural microbial communities. Use of the 16S rRNA Molecule Nucleic acid sequence comparisons have provided an invaluable perspective on the evolutionary relationships of microorganisms. Microbial systematics had previously relied on certain phenotypic characters of microorganisms, which were later found to be poor phylogenetic indicators. The most utilized molecule for nucleic acid sequence comparisons among microorganisms is the small subunit ribosomal RNA (16S rRNA). The 16S rRNA molecule is functionally and evolutionarily homologous among all organisms, due to its essential role in protein synthesis, and is not laterally transferred between microorganisms (Olsen et al., 1986). Thus, the phylogenetic relationships between the 16S rRNAs are reflective of the evolutionary relationships of the organisms. The sequence of the 16S rRNA molecule, or its gene, are mosaics of both highly conserved and variable regions, which extends the utility of this molecule to a wide phylogenetic range (Woese, 1987). 15 The utility of 16S rRNA sequence analysis has extended into the study of the ecology and diversity of natural populations of microorganisms. By linking the sequence analysis approach with current cloning technologies, a more complete view of the structure and composition of microbial communities is beginning to emerge. Novel 16S rRNA genes from bacterioplankton samples collected from the Sargasso Sea do not resemble genes from known bacterial species (Britschgi et al., 1991; Giovannoni et al., 1990). In addition, unique 16S rRNA genes belonging to the domain Archaea have recently been detected in the Pacific Ocean (Fuhrman et al., 1992). These analyses have also been extended to the analysis of microorganisms associated with marine snow (De Long et al., 1993) and to the examination of the endosymbiotic bacteria associated with the gutless bivalve Solemya reidi (Cary, 1994). These molecular techniques are ideally suited to analyze the microbial community associated with Alvinella pompejazza. Experimental Rationale The primary goal of this study is to characterize the evolutionary relationships of the predominant bacteria present in the epibiotic population of microorganisms associated with Alvinella pornpejana. The dominant member(s) of this population are filamentous bacteria. Due to the abundance of these bacteria in the population and the intimate association with A. pornpejana (see previous section), this bacterial form is believed to play an important symbiotic role. Therefore, its further characterization may be important in defining the nature of this symbiosis. In this study, cloning and sequencing methodologies were matched to genetically dissect the bacterial community associated with Alvinella 16 pompejana (Figure 3). A specimen of A. pompejana was collected from an active hydrothermal vent site at 13'N on the EPR and was preserved frozen to maintain the integrity of the nucleic acids. A small patch of bacteria was removed from the dorsal integument of the animal. DNA was purified from this sample. The 16S rRNA genes were amplified using the polymerase chain reaction (PCR) with general bacterial primers. These primers are known to hybridize to the 5' and 3' ends of the 16S rRNA genes of all known bacteria. Thus, this provides a representation of the complete bacterial 16S rRNA genes present in an environmental DNA sample. These primers have proven successful in amplifying diverse assemblages of bacterial 16S rRNA genes from natural populations. The amplified 16S rRNA genes were subsequently cloned into a plasmid vector to separate and propagate the amplified genes. In order to identify unique phylotypes and determine their numerical distribution in the library, each cloned 16S rRNA gene was screened using restriction fragment length polymorphism (RFLP) analysis. Assuming the numerical dominance of the phylotypes in the library is reflective of the actual distribution in the natural community, representatives from the four most dominant clone types were chosen for complete gene sequencing and phylogenetic analysis. 17 EXPERIMENTAL APPROACH Alvinella pompejana Isolate bacteria Extract and purify DNA f Amplify 16S rRNA genes Construct clone library Group clones using RFLP analysis Sequence and phylogenetically analyze representatives of the numerically dominant clone types Figure 3. Schematic outline of the experimental strategy used for the phylogenetic analysis of the epibiotic microbial community associated with the hydrothermal vent polychaete Alvinella pompejana. 18 METHODS Sample Collection. Specimens of Alvinella pompejana were collected in October 1991 during the joint French-US HERO (Hydrothermal Environment Research Observatory) expeditions at the Elsa vent site 13°N (2620m) on the East Pacific Rise (12°48'N, 103°56'W). Individuals were collected from worm colonies along the walls of active chimneys using the mechanical arm of the Deep Sea Research Vessel Alvin. The specimens were kept cold (<4°C) in a thermally insulated container until surfacing. Once on board, the animals were immediately frozen in liquid nitrogen and kept at -80°C until needed. Bacterial Removal and DNA Purification. A large patch of bacteria, including a portion of worm integument, was removed from a frozen A. pompejana individual using sterile forceps. The bacterial sample was completely homogenized in 5M guanidinium isothiocyanate, 50mM Tris (pH 7.4), 25mM EDTA, 0.8% 2-mercaptoethanol, using a Dounce homogenizer. The homogenate volume was doubled with 50mM Tris-Cl (pH 8.0), 25mM EDTA, and centrifuged at 10,000 rpm in a tabletop centrifuge for 15 minutes at room temperature to remove cellular debris. Nucleic acids were precipitated with 0.8 volumes of 100% isopropanol at room temperature for 1 hour. The precipitate was collected by centrifugation at maximum speed in a tabletop centrifuge for 30 minutes, washed with ice cold 70% ethanol, resuspended in TE buffer (10mM Tris -Cl, 1mM EDTA pH 8.0). The extracted bulk nucleic acids were digested with proteinase K (a final concentration of 500µg /ml at 50°C for 1 hour). The 19 sample was then extracted twice with an equal volume of phenol/chloroform (4:1) and once with an equal volume of chloroform/isoamyl alcohol (24:1). For each extraction the phases were allowed to completely separate and the aqueous layer was retained. Nucleic acids were precipitated overnight with 0.1 volumes of 3M sodium acetate (pH 5.2) and 2 volumes of cold 100% ethanol at -20°C. The precipitate was collected following centrifugation as above, washed with 70% ethanol, and resuspended in TE. To remove contaminating RNA's the sample was treated with RNase (33 ng/1_11) for 10 minutes at 65°C, and phenol purified as above. The integrity of the DNA sample was determined by agarose gel electrophoresis through a 0.8% agarose gel in 1X TAE (40mM Tris-acetate, 1mM EDTA). The final DNA concentration was determined spectrophotometrically. Amplification of 16S rRNA Genes. Bacterial 16S rRNA genes were amplified from the environmental sample using the polymerase chain reaction (PCR) using two general bacterial 16S rRNA primers: EubB (5'-AGA GUU UGA UCM UGG CUC AG-3') and EubA (5'-ACR CCN ACC TAG TGG AGG AA-3'); (Giovannoni, 1991). PCR amplifications were performed on a Coy thermal cycler (Coy Corporation, Ann Arbor, MI). The reaction conditions were: 50mM KC1, 10mM Tris-HC1 (pH 8.4); 2.5mM MgC12; 0.2mM each dATP, dCTP, dGTP, dTTP; 0.21.1M of each amplification primer; 1Ong of purified template DNA; 2.5U Taq polymerase (Promega), in a total volume of 1004 Amplification profile conditions were 95°C for 1 min., 55°C for 1 min., and 72°C for 3 min. for 30 cycles. The resulting PCR products were ethanol precipitated and analyzed by gel electrophoresis as described above. 20 Construction and Screening of Clone Library. Two separate clone libraries were constructed from a single A. pompejana specimen. The amplified genes were cloned using a plasmid vector (pCRII, Invitrogen) specifically designed to accommodate PCR products that contain single non-template dependent additions of deoxyadenosines at the 3' ends of the DNA duplexes. The only significant difference between the two libraries was that in the first attempt the host cells were less competent and resulted in a low transformation efficiency. The clone libraries were constructed using the TA Cloning System version 1.3 (Invitrogen) following the manufacturer's instructions, with the following modifications. The ligation reaction was performed with PCR products obtained from a reaction with a reduced number of cycles (30) and that had been ethanol precipitated prior to ligation. The transformants were plated onto LB agar plates (10g tryptone, lOg NaC1, 5g yeast extract, 15g bacto­ agar/L, pH 7.0) containing the antibiotic kanamycin (50µg /ml) with 25111 of X- Gal (40mg/m1) spread evenly across the agar plates. White colonies were scored and picked from all available plates and confirmed by restreaking. Stab cultures of positive clones were made in LB agar for long-term storage. Plasmid DNA was prepared from each of the confirmed transformants using an alkaline lysis preparation protocol (Sambrook et al., 1989). Confirmed white colonies were grown in 5m1 of LB media containing kanamycin (50ug/m1) overnight at 37°C with vigorous shaking. Two milliliters of this culture was used in the plasmid preparation. The purified plasmids were resolved on a 1% agarose gel in IX TAE and visualized with ethidium bromide staining. A plasmid known to contain the correct size insert and one lacking an insert (blue) were used as size standards. 21 Transformant plasmid DNAs which co-migrated with the 1.5 kb-insert control were scored as having a full-length 16S rRNA gene. RFLP Analysis. In order to sort the clones into similar clone types, clones containing full-length inserts were subjected to RFLP analysis. The cloned insert was re- amplified using the same PCR conditions and the primers used previously to establish the library. This amplification step was required to generate a sufficient amount of isolated product for the restriction analysis. 1111 of a 1:10 dilution of the each of the alkaline lysis plasmid DNA preparations was used as the starting template. Each PCR product was digested using two restriction endonucleases, Hae III and Mbo I (Promega), each of which recognize a four-base pair restriction site. 5111 of each PCR reaction was digested with 2.5U of each enzyme in 0.5X restriction buffer (5mM Tris-C1, 5mM MgC12, 25mM NaC1, 0.5mM DTT pH 7.9) at 37°C for 2 hours. The final reaction volume was 10 111. The reaction was stopped by the addition of 0.5p1 of 500mM EDTA (pH 8.0). The entire reaction volume was electrophoresed through a 3% Nu Sieve agarose gel (FMC) in IX TAE using a standard 6X loading buffer (Sambrook et al., 1989). Nu Sieve agarose was used in these analyses because it has been specifically developed for resolving DNA products less than 1000 bp. A third tracking dye, orange G, was added to the mixture (0.35% w/v final concentration) to aid in the running consistency between gels. 22 Gene Sequencing Representative clones from each of the four numerically dominant clone families resolved by the RFLP analyses were bidirectionally sequenced using a combination of both automated and manual plasmid sequencing. Automated sequencing was performed on an ABI model 373A automated gene sequencer at the Center for Gene Research and Biotechnology (Oregon State University). Manual sequencing was performed by standard dideoxynucleotide­ terminated sequencing protocols using a Sequenase v2.0 DNA sequencing kit (U.S. Biochemical Corp.) Sequencing reactions were performed following the supplier's instructions. Template plasmid DNA was prepared using Magic Mini plasmid purification kits (Promega). Two fig of purified DNA were denatured for each sequencing reaction. Template was denatured as follows: 2 lig of plasmid DNA were brought to a final concentration of 0.18M Na0H/0.18mM EDTA and incubated for 5 min. at room temperature. The DNA was precipitated with 0.1 volumes of 3M sodium acetate and 2 volumes of ice cold 100% ethanol for 30 min. at -70°C. The precipitate was collected by centrifugation at 20,000 rpm at 4°C for 30 min. in a Beckman TL-100 refrigerated tabletop ultracentrifuge. The pellet was washed in ice cold 70% ethanol and centrifuged. The pellet was allowed to air dry and was stored at -70°C until used for sequencing. Sequencing reactions were resolved by electrophoresis through a 7% polyacrylamide gel using Long-Ranger acrylamide (AT Biochem). 23 Phylogenetic Analysis The alvinellid clone 16S rDNA sequences were manually aligned to a subset of other aligned bacterial 16S rRNA sequences obtained from the Ribosomal Database Project (RDP) (Larsen et al., 1993). In addition, the 16S rRNA sequence for Desulfurella ncetivorans (EMBL accession number X72768) was aligned and included in the analysis. The conserved regions and established secondary structural models of the 16S rRNA sequence were used as guides to insure a correct alignment of the homologous regions of the sequences. Sequence data were manipulated using the Genetic Data Environment (GDE) v2.0 sequence analysis software package (Smith et al., 1992). Regions of ambiguous alignment i.e. positions 1-11, 69-102, 128-132, 176-226, 265-269, 451-480, 840-869, 999-1045, 1124-1185, 1262-1298, 1357-3' terminus (using the E. co/i numbering system) were excluded from the analysis. A total of 1061 nucleotide positions were used in the analysis. Phylogenetic trees were edited using the Treetools program (provided by Mike Maciukenas from the University of Illinois at Urbana-Champaign for the RDP) Programs used to infer phylogenetic trees are contained in the Phylip v3.5c software package (Felsenstein, 1993). DNADIST was used to calculate evolutionary distances with the Kimura 2-parameter model for nucleotide change and a transition/transversion ratio of 2.0 (Kimura, 1980). Phylogenetic trees were reconstructed from evolutionary distance data using the neighbor-joining method (Saitou et al., 1987), implemented through the program NEIGHBOR. Parsimony trees were reconstructed using DNAPARS. A total of 100 bootstrapped replicate resampling data sets for both DNADIST and DNAPARS were generated using SEQBOOT, with random sequence addition and global rearrangement. Secondary structural models for the 24 cloned 16S rDNA sequences were obtained with the gRNAID v1.4 program (provided by Shannon Whitmore) and optimized by comparison with other established 16S rRNA secondary structures (Gutell, 1993). 25 RESULTS Clone Library Construction DNA was isolated from a sample of the epibiotic microflora obtained from an A. pompejana individual and the bacterial 16S rRNA genes were amplified from this preparation. Two separate attempts were made at cloning these amplified genes. Both attempts used the TA Cloning System v1.3 (Invitrogen); however the first attempt utilized a version in which the competency of the cells was significantly lower. In the earlier attempt a total of 40 white transformants were obtained and all were screened for correct sized (1.5kb) inserts. Only 4 transformants was found to contain full-length inserts. The second attempt using more competent host cells resulted in 249 total white transformants. A total of 162 white transformants was screened and 135 of these contained the correct size inserts. Clones containing full- length inserts from both attempts were grouped into one library resulting in a final total of 139 clones. Clones from the first attempt are designated "APG #A", while those from the second library are designated "APG #B". Restriction Fragment Length Polymorphism Analysis In order to identify unique clone types, clones confirmed to contain a full-length insert were subjected to RFLP analysis. The insert was first amplified from purified plasmid DNA and the resulting amplification product was digested with two restriction endonucleases, Hae III and Mbo I. The clones were then grouped into "families" based on their restriction pattern; that is, clones with identical restriction patterns were placed into the 26 same family. The restriction analysis of the 139 full-length clones identified 32 distinct clone families. Figure 4 is a schematic representation of the restriction patterns of 11 representative clone families. This figure illustrates some of the diversity of restriction patterns seen in the library. Table 2 shows the numerical distribution of each of the clone families. Four of these families, designated Fl, F2, F9, and F13, dominated the library, representing 16.4% (23 clones), 43.2% (60 clones), 5.8% (8 clones), and 4.3% (6 clones) of the library, respectively. The remaining families were represented by fewer members, with the majority of the families containing only one member. Based on the assumption that the numerical dominance of the bacterial species will be reflected in the clone library, representative clones from the four most dominant families (Fl, F2, F9, and F13) were chosen for complete sequencing of the cloned insert. Because there is no evidence at this time to support this assumption, it has merely served as a starting point in the analysis of the microbial population associated with A. pompejana. The clones representing these four families are as follows: APG 5A (F1), APG 13B (F2), APG 56B (F9), and APG 44B (F13). 16S rRNA Gene Sequences The 16S rRNA gene sequences for the four alvinellid clones have been deposited in Gen Bank under the following accession numbers: APG 5A, L35523; APG 13B, L35520; APG 44B, L35521; and APG 56B, L35522. 27 M 1 Fl F2 1 1 F4 F9 1 1 111 III F10 F11 F13 F16 F17 F19 F20 M I 1018 1000 800 600 506 500 396 400 344 298 220 300 200 201 154 134 100 75 Figure 4. Scaled illustration of the restriction patterns of 16S rDNAs representing 11 clone families identified in the alvinellid done library. Each of these families contain 2 or more representative clones. Molecular weight standards (M) are included for comparison. The fragment sizes are given in base pairs. 28 Table 2. RFLP analysis results showing the numerical distribution of the alvinellid clone families. Clone Family Number of Clones ,t," % of Library 23 F F3 F4 F5 F6 F7 F8 F14 F15 F16 F 17 1 3 1 1 1 1 2 3 F18 1 F 19 F 20 5 2 F21 F22 1 F 23 F 24 F 25 F 26 F 27 F 28 F 29 F 30 F 31 F 32 1 1 1 1 1 1 1 1 0.7 2.2 0.7 0.7 0.7 0.7 0.7 1.4 2.2 0.7 3.6 1.4 0.7 0.7 0.7 0.7 0.7 0.7 0.7 0.7 0.7 0.7 0.7 0.7 a Highlighted rows indicate the four numerically dominant clone families chosen for phylogenetic characterization. 29 Phylogenetic Analysis Three different types of analyses were performed to determine the correct phylogenetic placement of the four alvinellid clones: 1) phylogenetic tree reconstruction inferred from both evolutionary distance and character based calculations, 2) comparison of secondary structural models of the clone 16S rRNA to other established 16S rRNA secondary structures, and 3) comparison of established signature base positions in the 16S rRNA molecule. Figure 5 is a phylogenetic tree inferred by the neighbor-joining method of phylogenetic tree reconstruction. This analysis includes the four alvinellid clone 16S rRNA sequences, a subset of reference sequences from the RDP, and the 16S rRNA sequence from a newly characterized bacterium, Desulfurella acetivorans (Rainey et al., 1993). Bootstrap values corresponding to 100 replicate re-samplings are shown above the lines leading to each branch, unless otherwise designated. This analysis included a total of 1061 nucleotide positions. The four alvinellid clone 16S rRNA sequences form their own cluster within the epsilon subdivision of the Proteobacteria. The alvinellid clones cluster together to the exclusion of other species, supported by a bootstrap value of 100%. This cluster clearly groups within the epsilon Proteobacteria, which is also supported by a bootstrap value of 100%. An identical tree topology was also obtained with the parsimony method, with bootstrap values of 99% and 100%, respectively, showing the robustness of these two branches (data not shown). Clones APG 5A and APG 13B are more closely related within this Glade, as are APG 44B and APG 56B. 30 Myxococcus xanthus o Desulfuromonas acetoxidans Desulfovibrio desulfuricans Flexispira rappini Helicobacter pylori Wolinella succinogens Arcobacter cryaerophilus 88 100 100 Campylobacter jejuni E Thiovulum sp. APG 5A APG 13B APG 44B APG 56B 100 100 100 78 Desulfurella acetivorans 100 87 100 98 79 95 Rhodospirillum rubrum Agrobacterium tumifaciens Nitrosomonas europae Pseudomcmas testosteroni Thiobacillus hydrothermalis a Escherichia coli 93 100 J Bathymodiolus thermophilus symbiont Calyptogena rnagmfica symbiont Thiomicrospira L-12 0.10 Figure 5. Phylogenetic tree showing the relationship of the alvinellid clones to the other members of the Proteobacteria. This tree was inferred from 16S rRNA sequence data using the neighbor-joining method. Molecular sequences for all reference strains (exceptDesulfurella acetivorans) was obtained from the RDP. Boldface type indicates 16S rRNA sequences cloned from the epibiotic microbial population of an Alvinella pornpejana individual. A total of 1084 nucleotide positions were included in the analysis. The tree was rooted with the sequence of Bacillus subtilis. Scale bar indicates 0.1 fixed mutations per nucleotide position. Numbers refer to boostrap values for each node out of a total of 100 replicate resamplings (values below 75 are not shown). 31 Despite the high confidence placed in the branch associating the alvinellid cluster with the epsilon Proteobacteria, the position of the alvinellid cluster within this subdivision is not supported as strongly. In the tree shown in Fig. 5, the alvinellid cluster appears as a sister group to the Campylobacter /Thiovulum group. This branch is supported by bootstrap values of only 57% by the neighbor-joining method, and 66% by the parsimony method. Examination of the alternative branching orders obtained with these analyses showed the alvinellid cluster varied in its position within the epsilon subdivision. The alvinellid cluster also appeared as a deeper group to the epsilon Proteobacteria (data not show). This variation was dependent on the nucleotide positions included in the analyses; however, the affiliation of the alvinellid cluster with the epsilon Proteobacteria was consistently recovered. Higher-order structural features of the 16S rRNA molecule are useful in the phylogenetic characterization of microbial groups (Woese, 1987b). Comparison of the 16S rRNA secondary structural models between distantly related groups, i.e. at the phyla and subdivision levels, can reveal structural features which support their phylogenetic relatedness. Comparison of these structural features between closely related groups is usually not as informative. Because 16S rRNA sequences for the alvinellid clones APG 5A/APG 13B and APG 44B/APG 56B are very similar it would be unlikely that reconstruction of a 16S rRNA secondary structural model for all four clone sequences would reveal significant defining characters. For this reason secondary structural models for only two of the alvinellid clones were generated. The alvinellid clones APG 13B and APG 56B were chosen to represent their respective groups. 32 Figures 6 and 7 show the proposed secondary structures of two alvinellid clone sequences, APG 13B and APG 56B. Both of these structures are consistent with the general topological features of other established bacterial 16S rRNA secondary structures. Several structural features were observed which support the placement of the alvinellid clones within the epsilon Proteobacteria. Three secondary structural features characteristic of the epsilon Proteobacteria (then referred to as the Campylobacter Thiovulum subdivision) had been previously reported (Lane et al., 1992). All three of these structural features have been observed in our alvinellid clones (position numbers are those of the 16S rRNA sequence of E. coli): 1) a conserved deletion of a helix corresponding to positions 455-477, 2) a unique compensatory base pair change at position 921 (U ) A) and 1396 (A U), and 3) a "long-short" stem arrangement involving stem structures at positions 184-193 and 198-219 respectively (Figure 6 and 7). In addition to these, another feature was observed which has been reported among the epsilon Proteobacteria (Rainey et al., 1993): an insertion of a (C) in a conserved loop structure corresponding to positions 1357-1365. This was observed independently of the discovery by Rainey and colleagues (1993). In order to confirm the results from the previous two phylogenetic analyses, an examination of the signature base positions distinguishing the subdivisions of the Proteobacteria was undertaken. Signature base positions are specific positions within the 16S rRNA molecule which have been used to distinguish various members of the bacterial, archace, and eukaryal domains (Woese, 1987). Because the epsilons are a newly recognized subdivision of the Proteobacteria, the signature base positions for these members have only recently been compiled (Rainey et al., 1993). In order to confirm the composition of these nucleotide positions, complete 16S rRNA 33 Figure 6. Proposed secondary structural model of clone APG 13B 16S rRNA. Numbers indicate regions which are discussed in text. This structural model was obtained using the gRNAID v 1.4 program and comparative analysis with other bacterial 16S rRNA structures (Gutell 1993). * D V V--V v-V 4-* <V-.V 40__ V-V 4 I, V 7-1 ii. D t ..--4 V 4-7VV 4V._ P-'" V44 ill%4 0444 1.V4V4/,',.,. V4,1,,,LI V. "ii V.-V v'i., 4 V4 13-13 v44 "D Ylit i 1Tr 1 t:tYYj 7y :",:V.v V, 4.p; ;::.1.1,,,, ;6\ 0i4;l: ::"rr,,:-.),.. . ,...;,5' ; V; 4,44 '-g =p V .,, . . v,,, v,,,, u=3V l3,1' %VD .V' =g 4 . 44 Vi.% '1,.(% v'\ " o' '4 " v3i tiv. 1=3 \ b-u 3=8 V-.0 D-4 V-V u-u t4" .-.1 .. u ": n' j.; 44.,, 7":: '' myD V""..IYT i S,,4r.-t ,J4 .-40y.V7V V" 4 .4..40 V4v. .,:::4 "v7 ',V 'f-(/V< 1 ../.>. ,, 4 v v D' ' "V IV 1, 41.1VivT,, TvInall,Yv %=4, ., U vVV 4,4 04V 40V 40 4di, I;4 4 0 .1313.-10 V. 0c.,V ' :04? 74VDV 44 4 7V4VV0VVOD.4 in * ,.u, 1), 44 ":''?::"1JriritYV v 4VV4 Dv V 40D V NI'-' "V 4,,0-IVO,' 0 LI41711 V44 V .tg D 74 LI...04-En D "4 ,,.0 0 13' '13 V-13 V ,, . 449. D' 4-.* D- -. '-'' V-V .0'4-10 V v . -... . -v _ , , V ''''' `'' ll'' uvuuvov u.yvvvpuv"...\.... Nu 01111.....Vitt -uu VO DV 0 04,V...f., u V /A V"( le ((;)v v . EMIL! Irati Vti.AIA, 1 l? V,v,, 4V" 0-,4 V D pA4Ei V-V \,..il. 0 ,.,0-.4., 44V 4--g - ulasb:4. o4 u° '1* 4 o r6­ a v 0 44 b 0 .....0 P-4 ° 4e 3 ' 4vu° 4 t;''V ', .0 V V BS _, r., : 'V .. 0 .u ' .4e o o4_ -V 3 0 V <<;V 41tWg4 V ....D., V ,--,k,.., 13 %Vv., .''' VL&-ki. 94 4 4 4 u v u V: 04 ". <-0 DO V .V v D .< v v 0'02 .11 4113-,3, i 7,YJRZ:740/E ,,,,.. 4-1 "-."4 44\*4V g UAW/ VV.., 31) vi.1.44V44VV 77tV VV4-, D 4...0 4..0 u u" is ,,vO' :...'--:' 4 4 3 1 *v 9, .1111TT:rf In1.4/Tv. 4 4.4 V 110 ,.,.1 04 44 4'...°..44" 4 ,....,..VV 0.40 4-'-'6 ` V" II-4 . 0 "70-4:13V 40"' .D-4 -i-i. ,; .°* U**Up 4 14 4/V.V:. '//". V Da ).., ... 35 Figure 7. Proposed secondary structural model of clone APG 56B 16S rRNA. Numbers indicate regions which are discussed in text. Positions with asterisks denote signature nucleotide' discrepancies which were further examined for compensatory base pair changes. This structural model was obtained using the gRNAID v 1.4 program and comparative analysis with other bacterial 16S rRNA structures (Gutell 1993). . V' 0-.4 D-4 4 .7 v 4.-0 v y_v 4 Do V .14 YYYYY V. V V -' V :'. la , ,,,,, b CJ b gt' ,4* ,.,DUV . 4V * v v.' 4 D-..4 1 ° v/4 U 4 U 0 D 4 v i.v.v v° .4 ° . 117B 8.v .4_5 V.. ...V I 4V V i.:q. 4.1.44/4.0....D.4..,-) .4 .V". %.t bioc ' <i 4 u ,,k\ "/.:/.. 4.- i.'? ././ `.' V V.-ty v_. 4-D ,,__. .... % D . bp u-<4- 1.11 , t(1 vUV. Ei '4. 4.,..) El ' ''.4 .4. 54 4 :t:trat Iti ,r DV ,Y-.? ..,..,,,, 4..,? . .,,,,, '1*.ice'D% :a u;.4 '' .4 < cs-to u e,ts v6..., Vo A 0.--U 4-D °"7- 44° V .%.:-,.. D. . ReD4'. V.,. AA,,, 1,<P'1, ..v ..IrttcEz As4; ,..._1.4,&,,,,ttiltki4r1 V° ,-\;..,, 4 /...,...c.,..... V ' It'Z ,14v ) *17:12* vUs.8 1.. .t,,,..._ . .V4Y g v. ,/,. '//". L(I/4 s,.....,,, 1,vu ' ':k:' ' 4 04 V t 4 '\42:141114VVIVVVDDV:tailEE/V.:itEitUiFIllit. D -1" 04., r-1 4,, V't.\74 V ° 4 0, itUIMIt, A VVV \ . b. ' ' '4-1?:' ' ' °, .7.,1, ..; L. t, :" vEintgUtT4LX.4I 4u olv .v....4 D'''VlUI:):f 4-. 4 .--.. tt . 4:::..,./ tk..;-4 <::Utt.,:''°,011it/i .4 4. ... * V.(,K;".,.. 0 4.., '''' %:!;\`'4\ '"` v_v ,..,. ..v. ejt,',' iv \'' i YYVVYY "'"V 04 V 04 V V. ..C,'4qtrkt aVTC E;''t Il tttai: ,',7 YD VD VV '4 '''''''' v 4 I, 1,;L:,1<bv v v ,4 V-V .-4 4i% us. via . v ..., Ct74 1,--10-ll-u x46 V, gD 44 V ..-<k*ov`v : !A11,tr ; ' vv ttHEU U.4 V ._. ., 40 .....0 'VV.,' 4.-0 Da .Y: V ..,,,, v-u .. 4-17 v-U .._.. v._ v ,,. ?v,,,, "A ... Vf <(,,... . I ,, - ­ ItitrEtat. ,,, 37 sequences representing 51 available epsilon Proteobacteria obtained from the RDP were examined. The signature base positions for the four alvinellid clones were compiled separately for comparison. Table 3 shows the signature base positions compiled for the epsilon and delta Proteobacteria. A total of 72 signature positions were examined. Out of these 72 positions, 48 were found to differ between the delta and epsilon subdivisions. These signature base positions were also examined in the alvinellid clones. Out of the 48 positions distinguishing the deltas from the epsilons, only four were found to differ between the epsilons and the alvinellid clones (see highlighted positions in Table 3). In order to rule out the possibility that these differences were due to experimental artifacts, these same positions were re-examined in the clone 16S rRNA secondary structural models. The discrepancies at positions 370, 391, and 640 were seen in two of the alvinellid clones, APG 44B and APG 56B. Secondary structure analysis for clone APG 56B supports these three discrepancies, revealing compensatory base pair changes at the adjoining positions in the stem structures of the molecule (Figure 7). Compensatory base pair changes are changes in variable nucleotides which preserve the conserved secondary structure of the 16S rRNA molecule(Gutell et al., 1990). This type of comparison is not possible with the difference at position 108 because it occurs in a non-stem structure. Table g. Signature base positions distinguishing the delta and epsilon subclasses of the Proteobacteria a and the alvinellid clones. Position Delta 6 Gb Epsilon C C C 7 44 A A 50 APG clones N. D. N. D. 107 .. 124 129 129:1 233 236 237 242 284 ..37. C A C 871 A:u A A C C C C C U C U A Y C C C:g G:c U U A A C:R:u G:a A U U:c A U U Ig.V.PagERIFAPPIRIEBRE 390 C:u g:a A:c:u Vn 689 690 698 722 760 823 825 kg U a 564 812 822 875 877 878 916 929 947 948 976 1015 1024 1026 1116 1120 1153 1219 1233 1234 1246 1252 1260 . 371 398 438 449 485 496 502 513 538 543 554 Position G:u A C C:a U:c U:a U:c U A U A C C A C C Delta C R:u R:u A:g U U:c Epsilon APG Clones C C Y:a Y:a G G U A:U:c:g G:u Y C G GA A:g G:a C A:U:g:c A R U Y R A C U C U G Y G A A G G C G G:a C C C C N Y:A R:U A:C U:G A R R R C:a C C G:c G Y Y:C G:u G:u R C C G A A:U A:u C:Y kg 1291 C:g Y 1297 1298 1325 Y C:A C Y 1421 Y C C:g A:g C U C:a C U U C G 1426 U:R C 1431 Y:a U U Y C C C C:g:a A U:g A 1437 1441 1443 G:u C A A R C The information for the delta subclass was taken from Table 4 of Woese (1987) and Table I of Rainey et al (1993). The information for the epsilon subclass was compiled from an alignment of 51 16S rRNA sequences of epsilon Proteobacterla. Y, pyrimidine; R, purine; N, any nucleotide; N. D., not determined. b Upper case, major base; if no other specified, it accounts for >90% of assayable cases. Lower case, minor occurrence base; found In <15% of assayable cases (or in only one sequence). c Shaded positions indicate nucleotide differences between the epsilon Proteobacteria and the alvinellid clones. 39 DISCUSSION Bacterial/invertebrate symbioses are well recognized in marine systems. However, defining the fidelity of these relationships is frequently problematic. Differentiating relationships that are commensal, where only one partner gains benefit from the relationship, from those that are truly mutualistic, where both partners profit, is difficult. The partners are usually so intricately tied that resolving the level of interaction has been impossible. Although considerably more is known about monospecific endosymbiotic associations in certain marine invertebrates (Cavanaugh, 1985; Fisher, 1990), there are numerous cases of diverse microbial assemblages, found obligately associated with the external surfaces of certain invertebrates, which have eluded sufficient explanation (de Burgh & Sing la, 1984; Johnson et al., 1981; Richards et al., 1982). Molecular genetic techniques now provide an avenue by which to study these tightly associated symbioses. The relationship between the hydrothermal vent polychaetes Alvinella spp. and their associated microflora has intrigued investigators since the discovery of these unique organisms (Desbruyeres & Laubier, 1980b). Both A. pornpejana and A. caudata possess morphologically and metabolically diverse assemblages of microorganisms associated with the worms' dorsal surfaces, some of which appear specifically attached (Gaill et al., 1987; Jeanthon & Prieur, 1990; Prieur et al., 1990b; Prieur et al., 1987a); Desbruyeres, 1985 #1636. It has been suggested that the dominant integrated morphotype, a filament, plays a significant role in the either the nutrition of its host or in the detoxification of the worms' immediate environment (Gaill et al., 1988). The primary focus of this study was to examine the diversity of 40 this unique microflora and, through the approach presented here, phylogenetically characterize several of the dominant phylotypes. The initial RFLP analysis of the alvinellid clone library grouped the 139 clones into similar clone types or families. The analysis identified 32 distinct clone families. Four of these clone families collectively represented over 70% of the total number of clones, with the remaining families consisting of only one or a few members. There is no evidence to suggest that the assumption that the distribution of clone types in the library represents the actual distribution in the natural population. However, the numerical distribution of the dominant clone families is in agreement with previous electron microscopic observations, which demonstrated that a filamentous morphotype dominated the microbial biomass (Gaill et al., 1987). The choice of clone types to phylogenetically characterize was based on their numerical dominance in the library. The 16S rRNA sequences of individual alvinellid clones representing the four dominant clone families and a subset of other representative bacterial 16S rRNA sequences were used to infer their evolutionary relationships. Both parsimony and distance phylogenetic inference methods placed the four alvinellid clones as a unique cluster within the epsilon Proteobacteria. Both methods gave a bootstrap value of 100%, illustrating the robustness of this placement. In the phylogenetic tree presented here (Figure 5) the alvinellid cluster is shown as a sister lineage to the Campylobacter/Thiovulum Glade. However, this association is not strongly supported; the corresponding bootstrap values are 57% and 66%, obtained with the neighbor-joining and parsimony methods, respectively. The next most strongly supported placement of the alvinellid cluster is as a deeper branching lineage within the epsilon subdivision. It is clear that the alvinellid clones form their own cluster within the epsilon 41 Proteobacteria to the exclusion of other species; however this analysis is not able to resolve the precise placement of the cluster within the epsilon subdivision with a high degree of confidence. Two additional comparative analyses of 16S rRNA sequences are commonly used to provide support for the phylogenetic affiliations of organisms: 1) the comparison of specific structural features in 16S rRNA secondary structural models and 2) the identification of specific signature base positions in the 16S rRNA sequence (Woese, 1987b). Both of these analyses were carried out with the sequences of the alvinellid clones, in order to lend additional support to the phylogenetic position obtained with the tree inference methods. The comparison of completed secondary structural models for two of the alvinellid clone 16S rRNAs with other established bacterial 16S rRNA secondary structures revealed several features that supported the inclusion of the alvinellid clones within the epsilon Proteobacteria. Lane and colleagues (1992), reported three important structural features which were conserved among several members of the epsilon subdivision (then referred to as the Thiovulum-Campylobacter subdivision). All three of these structural features were present in the alvinellid clones. In addition, an insertion of a cytosine residue in the conserved loop structure corresponding to nucleotide positions 1357-1363 (E. coil numbering system) was found in all of the alvinellid clones. The significance of this insertion was first reported in Desulfurella acetivorans, a more distant relative of the epsilon Proteobacteria (Rainey et al., 1993). A thorough analysis of other members of the epsilon subdivision supports this insertion as being highly conserved. Examination of both the 16S rRNA secondary structure of another epsilon proteobacterial member, Campylobacter sputorurn subsp. sputorum (Gutell, 1993). and of a 42 general alignment of epsilon 16S rRNA sequences obtained from the RDP suggests an equivalent insertion at this position. Examination of other proteobacterial 16S rRNA sequences showed that an insertion at this position occurs in only 10 out of 925 available 16S rRNA sequences from the four other established proteobacterial subdivisions. Thus, this insertion has the potential to become a defining character of the epsilon Proteobacteria. The presence of certain 16S rRNA signature nucleotides of the Proteobacteria (Woese, 1987a) provides the final criterion with which to evaluate the phylogenetic position of an organism within the phylum. Because the epsilons are a newly recognized member of the Proteobacteria, the signature base positions for this group have only recently been compiled (Rainey et al., 1993). A total of 72 positions were examined in 51 available epsilon 16S rRNA sequences from the RDP. Comparison of these signatures to the alvinellid clones was consistent with their placement in the epsilon Proteobacteria. Only four positions were found to differ significantly between the alvinellid clones and the epsilons, and at least three of these differences appear to be real characteristics of the clone sequences. In each of these cases the nucleotide differences were supported by compensatory base pair changes in adjoining nucleotide positions of the clone secondary structure in which they occur. This type of analysis has been used previously to rule out nucleotide differences which might be due to sequencing errors or PCR artifacts (De Long et al., 1993). Recently a newly isolated bacterium, Desulfurella acetivorans, was phylogenetically characterized as a distant relative to the epsilon Proteobacteria, possibly representing a new subclass of this phylum (Rainey et al., 1993). These investigators used phylogenetic tree reconstruction and comparison of signature nucleotides of 16S rRNA sequences to infer the 43 evolutionary relationship of this thermophilic sulfur-reducing bacterium. The analysis presented in the current study confirms the phylogenetic position of D. acetivorans proposed by Rainey and co-workers; however, it has identified several discrepancies between a number of the key signature nucleotides for the epsilon Proteobacteria used in their analysis (data not shown). Although it is not possible to resolve these inconsistencies at this time, further examination of additional epsilon representatives will more firmly establish the epsilon nucleotide signatures for use in future studies. It is often difficult to infer the conserved phenotypic characters of a particular group of microorganisms based solely on their phylogenetic relatedness. However, phenotypic qualities that appear in ancestral branches and within more closely related species may provide some insight into the metabolic potential and ecology of unknown but related organisms. Currently there are seven known genera which form the two clusters comprising the epsilon subdivision of the Proteobacteria (Eaton et al., 1993; Lau et al., 1987; Lee et al., 1992; Paster et al., 1988; Paster et al., 1991; Romaniuk et al., 1987; Sly et al., 1993; Thompson et al., 1988; Vandamme et al., 1991) . A survey of the conserved phenotypic characters among these epsilon genera reveals that the majority are adapted to environments low in oxygen; they range from microaerophiles to some which are able to grow anaerobically (Holt et al., 1994; Kreig et al., 1984; Vandamme et al., 1991) . In addition, all of these genera, except the sulfur-oxidizing marine bacterium Thiovulum sp. have been found associated with a eukaryotic organism as a host, occurring in digestive tracts, reproductive organs, or oral cavities of higher vertebrates. These conserved phenotypic characters are consistent with the environment of the alvinellid epibionts. As well as being associated with a metazoan host, 44 these bacteria would also be adapted to the hypoxic conditions within the tubes of A. pompejana (Chevaldonne et al., 1991). In conclusion, the novel molecular approach described here has allowed the assessment of the microbial diversity of the epibiotic bacterial population associated with A. pompejana. From an established 16S rRNA clone library containing 32 distinct clone variants, several dominant phylotypes were characterized as members of the epsilon subdivision of the Proteobacteria. Future studies are currently underway to design and test oligodeoxynucleo tide probes specifically targeting regions of the Alvinella cloned 16S rRNA's. Previous studies have established the utility of such probes in identifying the morphology of a cell type through in situ hybridization techniques (Amann et al., 1991; Amann et al., 1990; Reysenbach et al., 1994); Giovannoni, 1988 #533. These experiments will determine the morphology of the clone types. The probes can then be used to screen large insert DNA libraries established directly from epibiont nucleic acid samples. This approach has been applied before to locate functional genes in free-living marine archebacteria (Stein, 1994). The functional genes can then be related to specific clone types, contributing additional information about the metabolic potential of these epibionts. Through these molecular approaches the function of the relationship between the Alvinella spp. and their associated bacterial population can be determined without the need to culture the organisms. 45 BIBLIOGRAPHY Alayse-Danet, A.-M., F. Gaill, & D. Desbruyeres (1986). In situ carbonate uptake by bacteria-Alvinella associations. Mar. Ecol., 7, 233-240. Alayse-Danet, A. M., D. Desbruyeres, & F. Gail (1987). The possible nutritional or detoxification role of the epibiotic bacteria of Alvinellid polychaetes: review of current data. Symbiosis, 4, 51-62. Amann, R., N. Springer, W. Ludwig, H. Gortz, & K. Schleifer (1991). Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature, 351, 161-164. Amann, R. I., L. Krumholz, & D. A. Stahl (1990). Fluorescent oligonucleotide probing of whole cells for determinative, phylogene tic and environmental studies in microbiology. J. Bacteriol., 172, 762-770. Arp, A. J., & J. J. Childress (1983). Sulfide binding by the blood of the hydrothermal vent vestimentiferan tube worm Riftia pachyptila. Science, 219, 295-297. Bodammer, J. E., & T. K. Sawyer (1981). Aufwuchs protozoa and bacteria on the gills of the rock crab, Cancer irroratus say: a survey by light and electron microscopy. T. Protozool., 28(1), 35-46. Britschgi, T. R., & S. J. Giovannoni (1991). Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Appl. Env. Microbiol., 57(6), 1707-1713. Cary, C. (1994). Vertical transmission of a chemoautotrophic symbiont in the proteobranch bivalve, Solemya reidi. Mol. Mar. Biol. Biotech., 3, 121-130. Cavanaugh, C. M. (1985). Symbiosis of chemolithotrophic bacteria and marine invertebrates from hydrothermal vents and reducing sediments. Biol. Soc. Wash. Bull., 6, 373-388. Cavanaugh, C. M., S. L. Gardiner, M. L. Jones, H. W. Jannasch, & J. B. Waterbury (1981). Procaryotic cells in the hydrothermal vent tube worm. Science, 213, 340-342. Chevaldonne, P., D. Desbruyeres, & J. J. Childress (1992). hotter. Nature, 359(6396), 593-594. ... and some even 46 Chevaldonne, P., D. Desbruyeres, & M. Le Haitre (1991). Time-series of temperture from three deep-sea hydrothermal vent sites. Deep Sea Res., 38, 1417-1430. Collins, Y. E., & G. Stotzky (1989). Factors affecting the toxicity of heavy metals to microbes. In T. J. Beveridge & R. J. Doyle (Eds.), Metal ions and bacteria (pp. 31-90). New York: John Wiley and Sons. De Burgh, M. E., & C. L. Singla (1984). Bacterial colonization and endocytosis on the gill of a new limpet species from a hydrothermal vent. Mar. Biol, 84, 1-6. DeLong, E. F., D. G. Franks, & A. L. Alldredge (1993). Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr., 38(5), 924-934. Desbruyeres, D., P. Crassous, J. F. Grassle, A. Khripounoff, D. Reyss, M. Rio, & M. Van Praet (1982). Donnees ecologiques sur un nouveau site d'hydrothermalisme actif de la ride du Pacifique oriental. C. R. Acad. Sci. Paris, ser. III, 295, 489-494. Desbruyeres, D., F. Gaill, L. Laubier, & Y. Fouquet (1985). Polychaetous annelids from hydrothermal vent ecosystems: An ecological overview. Bull. Biol. Soc. Wash., 6, 103-116. Desbruyeres, D., F. Gaill, L. Laubier, D. Prieur, & G. Rau (1983). Unusual nutrition of the "Pompeii worm" Alvinella pornpejana (polychaetous annelid) from a hydrothermal vent environment: SEM, TEM, 13C and 15N evidence. Mar. Biol., 75, 201-205. Desbruyeres, D., & L. Laubier (1980). Alvinella pornpejana gen. sp. now., aberrant Ampharetidae from East Pacific Rise hydrothermal vents. Oceanol. Acta, 3(3), 267-274. Desbruyeres, D., & L. Laubier (1986). Les Alvinellidae, une famille nouvelle d'annelides polychetes infeodees aux sources hydrothermales sous­ marines: systematique, biologie et ecologie. Can. T. Zool., 64, 2227-2245. Desbruyeres, D., & L. Laubier (1991). Systematics, phylogeny, ecology and distribution of the Alvinellidae (Polycheata) from deep-sea hydrothermal vents. Ophelia, 5, 31-45. 47 Durand, P., D. Prieur, C. Jeanthon, & E. Jacq (1990). Presence et actiyite de bacteries heterotrophes mangano-oxydantes associees aux Alvinellidae (Annelides polychetes) dans un site d'hydrothermalisme profond de la dorsale du Pacifique oriental. C. R. Acad. Sci. Paris, ser. III, 310, 273-278. Eaton, K. A., F. E. Dewhirst, M. J. Raclin, J. G. Fox, B. J. Paster, S. Krakowka, & D. R. Morgan (1993). Helicobacter acinonyx sp. nov., isolated from cheetas with gastritis. Int. T. Sys. Bacter., 43(1), 99-106. Felbeck, H. (1981). Chemoautotrophic potential of the hydrothermal vent tube worm, Riftia pachyptila (Vestimentifera). Science, 213, 336-338. Felsenstein, J. (1993). PHYLIP (Phylogeny Inference Package). Distributed by the author. Dept. of Genetics. U. of Washington, Seattle. Fisher, C. R. (1990). Chemoautotrophic and methanotrophic symbioses in marine invertebrates. Rev. Aquat. Sci., 2, 399-613. Fuhrman, J. A., K. McCallum, and A. A. Davis. (1992). Novel major archebacterial group from marine plankton. Nature (London) 356, 148­ 149. Gaill, F., D. Desbruyeres, & L. Laubier (1988). Relationships between the "Pompei worms" and their epibiotic bacteria. Oceanol. Acta, 8, 147-154. Gaill, F., D. Desbruyeres, & D. Prieur (1987). Bacterial communities associated with "Pompeii worms" from the East Pacific Rise hydrothermal vents: SEM, TEM observations. Microb. Ecol., 13, 129-139. Gaill, F., & S. Hunt (1986). Tubes of deep sea hydrothermal vent worms Riftia pachyptila (Vestimentifera) and Alvinella pornpejann (Annelida). Mar. Ecol. Prog. Ser., 34, 267-274. Giovannoni, S. J. (1991). The polymerase chain reaction. In E. Stackebrandt & M. Goodfellow (Eds.), Nucleic Acid Techniques in Bacterial Systematics (pp. 177-203). John Wiley & Sons Ltd. Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, & K. G. Field (1990). Genetic diversity in Sargasso Sea bacterioplankton. Nature (London), 345, 60-63. Gutell, R. R. (1993). Collection of small subunit (16S- and 16S-like) ribosomal RNA structures. Nucleic Acids Res., 21, 3051-3054. Gutell, R. R., & C. R. Woese (1990). Higher order structural elements in ribosomal RNAs: pseudo-knots and the use of noncanonical pairs. Proc. Natl. Acad. Sci., 87, 663-667. 48 Holt, J. G., N. R. Kreig, P. H. A. Sneath, J. T. Staley, & S. T. Williams (1994). Berey's manuel of determinative bacteriology (9th ed.). Baltimore: Williams and Wilkins. Jeanthon, C., T. Barbeyron, M. Mergeay, L. Die ls, & D. Prieur (1992). Plasmid­ determined resistance to arsenite in bacteria isolated from hydrothermal vents. Appl. Environ. Microbiol. Jeanthon, C., & D. Prieur (1990). Susceptibility to heavy metals and characterization of heterotrophic bacteria isolated from two hydrothermal vent polychaetes, Alvinella pompejana and Alvinella caudata. Appl. Environ. Microbiol., 56, 3308-3314. Johnson, I., N. Flower, & M. W. Loutit (1981). Conribution of periphytic bacteria to the concentration of chromium in the crab He lice crassa. Microb. Ecol., 7, 245-252. Kimura, M. (1980). A simple model for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. L Mol. Evol., 16, 111-120. Kreig, N. R., & J. G. Holt (Ed.). (1984). Bergey's manuel of systematic bacteriology. Baltimore, M.D.: Williams and Wilkins. Lane, D. L., A. P. Harrison, D. Stahl, B. Pace, S. J. Giovannoni, G. J. Olsen, & N. R. Pace (1992). Evolutionary relationships among sulfur-and ironoxidizing Eubacteria. J. Bac., 174(1), 269-278. Larsen, N., G. J. Olsen, B. L. Maidak, M. J. McCaughey, R. Overbeek, T. J. Macke, T. L. Marsh, & C. R. Woese (1993). The ribosomal database project. Nucleic Acids Res., 21 Suppl., 3021-3023. Lau, P. P., V. B. De Brunner, B. Dunn, K. Miotto, M. T. MacDonell, D. M. Rollins, C. J. Pillidge, R. B. Hespell, R. R. Colwell, & a. 1. et (1987). Phylogenetic diversity and position of the genus Campylobacter. Syst. Appl. Microbiol., 9(3), 231-8. Lee, A., M. W. Phillips, J. L. O'Rourke, B. J. Paster, F. E. Dewhirst, G. J. Fraser, J. G. Fox, L. I. Sly, P. J. Romaniuk, T. J. Trust, & S. Kouprach (1992). Helicobacter muridarum sp. nov., a microaerophillic helical bacterium with a novel ultrastructure isolated from the intestinal mucosa of rodents. Int. T. Sys. Bacteriol., 42(1), 27-36. McFall-Ngai, M. J., & E. G. Ruby (1991). Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science, 254, 1491-1494. 49 National Research Council. (1979). Hydrogen Sulfide. Baltimore: University Park Press. Olsen, G. J., D. J. Lane, S. J. Giovannoni, & N. R. Pace (1986). Microbial ecology and evolution: A ribosomal RNA approach. Annu. Rev. Microbiol., 40, 337-355. Paster, B. j., & F. E. Dewhirst (1988). Phylogeny of campylobacters, wolinellas, Bacteroides gracilis, and Bacteroides ureolyticus by 16S ribosomal ribonucleic acid sequencing . Int. T. Syst. Bacteriol., 38(1), 56-62. Paster, B. J., A. Lee, J. G. Fox, F. E. Dewhirst, L. A. Tordoff, G. J. Fraser, J. L. O'Rourke, N. S. Taylor, & R. Ferrero (1991). Phylogeny of Helicobacter felis sp. nov., Helicobacter mustelae, and related bacteria. Int. T. Sys. Bacter. , 41(1), 31-38. Powell, M. A., & G. N. Somero (1983). Blood components prevent sulfide poisoning of respiration of the hydrothermal vent tubeworm Riftia pachyptila. Science, 219, 297-299. Prieur, D., S. Chamroux, P. Durand, G. Erauso, P. Fera, C. Jeanthon, L. Le borgne, G. Mevel, & P. Vincent (1990). Metabolic diversity in epibiotic flora associated with the pompeii worms, Alvinella pompejana and Alvinella caudata (Polychaeta: Annelida) from deep-sea hydrothermal vents. Mar. Biol., 106, 361-367. Prieur, D., & C. Jeanthon (1987). Preliminary study of heterotrophic bacteria isolated from two deep sea hydrothermal vent invertebrates: Alvinella pornpejana (Polychaete) and Bathymodiolus thermophilus (Bivalve). Symbiosis, 4, 87-98. Rainey, F. A., R. Toalster, & E. Stackebrandt (1993). Desulfurella acetivorans, a thermophilic, acetate-oxidizing and sulfur-reducing organism, represents a distinct lineage within the Proteobacteria. System. Appl. Microbiol., 16, 373-379. Reysenbach, A., G. S. Wickham, & N. R. Pace (1994). Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl. Env. Micro., 60, 2113-2119. Richards, K. S., T. P. Fleming, & B. G. M. Jamieson (1982). An ultrastructural study of the distal epidermis and the occurrence of subcuticular bacteria in the gutless tubificid Phallodrilus albidus (Oligochaeta : Annelida). Aust. T. Zool., 30, 327-36. 50 Romaniuk, P. J., B. Zoltowska, T. T. Trust, D. J. Lane, G. J. Olsen, N. R. Pace, & D. A. Stahl (1987). Campylobater pylori, the spiral bacterium associated with human gastritis, is not a true Campylobacter sp. T. Bacteriol., 169, 2137-2141. Saitou, N., & M. Nei (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 4, 406-425. Sambrook, J., E. F. Fritsch, & T. Maniatis (1989). Molecular cloning: a laboratory manuel 2nd ed. Cold Spring Harbor, N.Y.: Cold Spring Harbor Lab. Press. Saulinier-Michel, C., F. Gaill, A. Hi ly, & P. Alberic (1990). Structure and functions of the digestive tract of Alvinella pompejana, a hydrothermal vent polychaete. Can. J. Zool., 68, 722-732. Silver, S., T. K. Misra, & R. A. Laddaga (1989). Bacterial resistance to toxic heavy metals. In T. J. Beveridge & R. J. Doyle (Eds.), Metal ions and bacteria New York: John Wiley and Sons. Sly, L. I., M. A. Bronsdon, J. P. Bowman, A. Holmes, & E. Stackebrandt (1993). The phylogenetic position of Helicobacter nemestrinae. Int. T. Sys. Bacter., 43(2), 386-387. Smith, D. C., & A. E. Douglas (1987). The biology of symbiosis. Baltimore: Edward Arnold. Smith, S. W., R. Overbeek, G. Olsen, C. Woese, P. M. Gillevet, & W. Gilbert (1992). Genetic data environment and the Harvard genome database. In Genome mapping and sequencing Cold Spring Harbor: Somero, G. N., J. F. Siebenaller, & P. W. Hochachka (1983). Biochemical and physiological adaptations of deep-sea animals. In G. T. Rowe (Eds.), Deepsea Biology (pp. 261-330). New York: John Wiley. Thompson, L. M. I., R. M. Smibert, J. L. Johnson, & N. R. Krieg (1988). Phylogenetic study of the genus Campylobacter. Int. J. Syst. Bacteriol., 38(2), 190-200. Toulmond, A., & F. E. I. Silitine (1990). Extracellular hemoglobins of hydrothermal vent annelids: structural and functional characteristics in three alvinellid species. Biol. Bull., 179, 366-373. 51 Tunnicliffe, V., D. Desbruyeres, D. Jollivet, & L. Laubier (1993). Systematic and ecological characteristics of Paralvinella sulfincola Desbruyeres and Laubier, a new polychaete (family Alvinellidae) from northeast Pacific hydrothermal vents. Can. T. Zool., 71, 286-297. Tunnicliffe, V., & S. K. Juniper (1990). Dynamic character of the hydrothermal vent habitat and the nature of sulphide chimney fauna. Prog. Oceanogr., 24, 1-13. Vandamme, P., E. Falsen, R. Rossau, B. Hoste, P. Segers, R. Tytgat, & J. De Ley (1991). Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int. T. Sys. Bacter., 41(1), 88-103. Vovelle, J., & F. Gaill (1986). Donnees morphologiques, histochimiques et microanalytiques sur l'elaboration du tube organomineral d'Alvinella pompejana, Polychete des sources hydrothermales, et leurs implications phylogenetiques. Zoologica Scripta, 15, 33-45. Woese, C. R. (1987). Bacterial Evolution. Microbiol. Rev., 51, 221-271. APPENDICES 52 APPENDIX 1 LOCUS APG5A 1496 by DNA 11-AUG-1994 DEFINITION Epibiont of Alvinella pompejana of 16S rRNA gene encoding 16S ribosomal RNA. KEYWORDS 16S ribosomal RNA. SOURCE Epibiont of Alvinella pompejana, 5A from APG. ORGANISM Epibiont of Alvinella pompejana Bacteria /Prokaryota; Proteobacteria; subgroup: epsilon. REFERENCE 1 (bases 8 to 1542) AUTHORS Haddad,M.A., Camacho,F. and Cary,S.C. TITLE Phylogenetic characterization of the epibiotic bacteria associated with the hydrothermal vent polychaete, Alvinella pompejana JOURNAL in prep, 0-0 (1994) STANDARD full automatic COMMENT Data kindly submitted in computer readable form by: S. Craig Cary Ph.D Microbiology Oregon State University Corvallis Oregon 97331 USA Phone: (503) 737-1837 Email: caryc@ava.bcc.orst.edu Fax: (503) 737-0496 [1] Author requested hold until 01-OCT-1994 Location/Qualifiers 1..1496 rRNA /gene="16S rRNA" iproduct="16S ribosomal RNA" FEATURES BASE COUNT 415 a 321 c 446 g 311 t 3 others ORIGIN 1 AGAGTTTGAT CCTGGCTCAG AGTGAACGCT GGCGGCGTGC TTAACACATG CAAGTCGAAC 61 GGTAACAGAT CTTCGGAGTG CTGACGAGTG GCGAACGGGT GAGTAAGGTA TAGCTAACTT 121 GCCTCTTGGA GAGGGATAGC CACTGGAAAG GGTGATTAAT ACCTCATACT CCTCTTAACT 181 ATAAGGTTAA GAGGGAAATG GTTTATTCCG CCAAGGGATA GGGCTATATG GTATCAGTTA 241 GTTGGTGGGG TAAAGGCCTA CCAAGACAAT GACGCCTAGC TGGTCTGAGA GGATGATCAG 53 301 CCACACTGGA ACTGAGACAC GGTCCAGACT CCTACGGGAG GCAGCAGTGG GGAATATTGC 361 ACAATGGGGG AAACCCTGAT GCAGCGACGC CGCGTGGAGG AAGAAACCCT TAGGGGCGT A 421 AACTCCTTTT ATAGTCGAAG AAGATGACGG TAGACTATGA ATAAGCACCG GCTAACTCCG 481 TGCCAGCAGC CGCGGTAATA CGGAGGGTGC AAGCGTTACT CGGAATCACT GGGCGTAAAG 541 CGCATGTAGG CGGTTAATTA AGTTAGAAGT GAAAGCCCAC AGCTTAACTG TGGAACAGCT 601 TCCAAAACTG ATTAACTAGA GTCTGGGAGA GGAAGATGGA ATTAGTAGTG TAGGGGTAAA 661 ATCCGTAGAG ATTACTAGGA ATACCGAAAG CGAAGGCGAT CTTCTGGAAC AGGACTGACG 721 CTGAGATGCG AAAGCGTGGG GAGCAAACAG GATTAGATAC CCTGGTAGTC CACGCCCTAA 781 ACGATGAATG TTAGTCGTTG GAGTGCTAGC CACTTCAGTG ATGCAGCTAA CGCATTAAAC 841 ATTCCGCCTG GGGAGTACGG CCGCAAGGTT AAAACTCAAA GGAATAGACG GGGACCCGCA 901 CAAGTGGTGG AGCATGTGGT TTAATTCGAA GATACGCGAA GAACCTTACC TGGCCTTGAC 961 ATATACCAGA ACCCACCAGA GATGGTGGGG TGCCTTCGGG AGCTGGTATA CAGGTGCTGC 1021 ACGGCTGTCG TCAGCTCGTG TCGTGAGATG TTGGGTTAAG TCCCACAACG AGCGCAACCC 1081 TCGTGACTAG TTACTAACAG CATAGGCTGA GGACTCTAGT CAGACTGCCT TCGTAAGGAG 1141 GAGGAAGGTG GGGACGACGT CAAGTCATCA TGGCCCTTAC GGCCAGGGCG ACACACGTGC 1201 TACAATGGGG AGGACAATGA GAAGCGAAAT CGCGGGATGG AGCAAATCTA TAAACCTTCT 1261 CTCAGTTCGG ATTGCAGTCT GCAACTCGAC TGCATGAAGC TGGAATCACT AGTAATCGTG 1321 AATCAGCCAT GTCACGGTGA ATACGTTCCC GGGTCTTGTA CTCACGCCCG TCACACCATG 1381 GGAGTTGATT TCACCCGAAG CGGGGATGCT AAATAGGCTA CCCGCCACGG TGGAAGACTG 1441 GGGTGAAGTC GTAACAAGGT AACCGTAGGA GNACCTGTGG NTGGATCACC NCCTTA 54 APPENDIX 2 LOCUS APG13b 1502 by rRNA 11-AUG-1994 DEFINITION Epibiont of Alvinella pompejana of 16S rRNA gene encoding 16S ribosomal RNA. KEYWORDS 16S ribosomal RNA. SOURCE Epibiont of Alvinella pompejana, 13B from APG. ORGANISM Epibiont of Alvinella pompejana Bacteria/Prokaryota; Proteobacteria; subgroup: epsilon. REFERENCE 1 (bases 8 to 1542) AUTHORS Haddad,M.A., Camacho,F. and Cary,S.C. TITLE Phylogenetic characterization of the epibiotic bacteria associated with the hydrothermal vent polychaete, Alvinella pompejana JOURNAL in prep, 0-0 (1994) STANDARD full automatic COMMENT Data kindly submitted in computer readable form by: S. Craig Cary Ph.D. Microbiology Oregon State University Corvallis Oregon 97331 USA Phone: (503) 737-1837 Email: caryc@ava.bcc.orst.edu Fax: (503) 737-0496 [1] Author requested hold until 01-OCT-1994 Location/Qualifiers rRNA 1..1502 /gene="16S rRNA" /product="16S ribosomal RNA" FEATURES BASE COUNT 424 a 319 c 437 g 320 t 2 others ORIGIN 1 AGAGTTTGAT CATGGCTCAG AGTGAACGCT GGCGGCGTGC TTAACACATG CAAGTCGAAC 61 GGTAACAGGT CTTCGGAGTG CTGACGAGTG GCGAACGGGT GAGTAATGTA TAGTTAATTT 121 TCCCCTTGGA GAGGGATAGC CACTGGAAAC GGTGATTAAT ACCTCATACT CCTTTTAACC 181 AAAAGGTTGA AAGGGAAATG GTTTATTCTG CCAAGGGATA AGACTATATC CTATCAGCTA 241 GTTGGTAGTG TAAGGGACTA CCAAGGCAAT GACGGGTAGC TGGTCTGAGA GGATGATCAG 55 301 CCACACTGGA ACTGAGACAC GGTCCAGACT CCTACGGGAG GCAGCAGTGG GGAATATTGC 361 ACAATGGGGG AAACCCTGAT GCAGCAACGC CGCGTGGAGG AAGAAGCATT TAGGTGTGTA 421 AACTCCTTTT ATCAAGGAAG AAGACGACGG TACTTGATGA ATAAGCACCG GCTAACTCCG 481 TGCCAGCAGC CGCGGTAATA CGGAGGGTGC AAGCGTTACT CGGAATCACT GGGCGTAAAG 541 CGCATGTAGG CGGTTGATTA GGCTCAACCG TAGAACAGCT 601 TCTAAAACTG ATTAACTAGA ATTAGTAGTG TAGGGGTAAA 661 ATCCGTAGAG ATTACTAGGA CTTCTGGAAC AGGACTGACG 721 CTGAGATGCG AAAGCGTGGG CCTGGTAGTC CACGCCCTAA AGTTAGAAGT GAAAGCCTAC GTCTGGGAGA GGAAGATGGA ATACCGAAAG CGAAGGCGAT GAGCAAACAG GATTAGATAC 781 ACGATGAATG 'TTAGTCGTTG GAGTGCTAGC CACTTCAGCG ATGCAGCTAA CGCATTAAAC 841 ATTCCGCCTG GGGAGTACGS CCGCAAGGTT AAAACTCAAA GGAATAGACG GGGACCCGCA 901 CAAGTGGTGG AGCATGTGGT TTAATTCGAA GATACGCGAA GAACCTTACC TGGCCTTGAC 961 ATATATCAGA ACCCACCAGA GATGGTGGGG TGCCTTCGGG AGCTGATATA CAGGTGCTGC 1021 ACGGCTGTCG TCAGCTCGTG TCGTGTGATG TTGGGTTAAG TCCCGCAACG AGCGCAACCC 1081 TCGTGACTAG TTACTAACAG CTTAGGCTGA GGACTCTAGT CAGACTGCCT TCGTAAGGAG 1141 GAGGAAGGTA AGGACGACGT CAAGTCATCA TGGCCCTTAC GGCCAGGGCG ACACACGTGC 1201 TACAATGGGA AGGACAAAGA GAAGCAAAAC TGCGAGGTGG AGYAAATCTA TAAACCTTCT 1261 CTCAGTTCGG ATTGCAGTCT GCAACTCGAC TGCATGAAGC TGGAATCACT AGTAATCGTG 1321 AATCAGCCAT GTCACGGTGA ATACGTTCCC GGGTCTTGTA CTCACCGCCC GTCACACCAT 1381 GGGAGTTGAT TTCACCCGAA GTGGGGATGC TAAATAGGCT ACCCACCACG GTGGAATTAG 1441 CGACTGGGGT GAAGTCGTAA CAAGGTAACC GTAGGAGAAC CTGCGGCTGG ATCACCTCCT 1501 TA 56 APPENDIX 3 LOCUS APG44b 1502 by DNA 11-AUG-1994 DEFINITION Epibiont of Alvinella pompejana of 16S rRNA gene encoding 16S ribosomal RNA. KEYWORDS 16S ribosomal RNA. SOURCE Epibiont of Alvinella pompejana, 5A from APG. ORGANISM Epibiont of Alvinella pompejana Bacteria / Prokaryota; Proteobacteria; subgroup: epsilon. REFERENCE 1 (bases 8 to 1542) AUTHORS Haddad,M.A., Camacho,F. and Cary,S.C. TITLE Phylogenetic characterization of the epibiotic bacteria associated with the hydrothermal vent polychaete, Alvinella pompejana JOURNAL in prep, 0-0 (1994) STANDARD full automatic COMMENT Data kindly submitted in computer readable form by: S. Craig Cary Ph.D Microbiology Oregon State University Corvallis Oregon 97331 USA Phone: (503) 737-1837 Email: caryc@ava.bcc.orst.edu Fax: (503) 737-0496 [1] Author requested hold until 01-OCT-1994 FEATURES Location/Qualifiers rRNA 1..1502 /gene="16S rRNA" /product="16S ribosomal RNA" BASE COUNT 399 a 344 c 449 g 310 t ORIGIN 1 AGAGTTTGAT CCTGGCTCAG AGTGAACGCT GGCGGCGTGC TTAACACATG CAAGTCGAAC 61 GGTAACAGCA GCTTGCACTG GGCTGACGAG TGGCGAACGG GTGAGTAGTG TATAGCTAAT 121 ATGCCCTTTG GAGGGGGATA GCCACTAGAA ATGGTGATTA ATACCCCATA CTACTTCTTC 181 TCACAAGAGT TGATGGGAAA TCTTTTTTGG CCAAAGGATT GGGCTATACG GTATCAGCTT 241 GTTGGTGAGG TAACTGCTCA CCAAGGCTAT GACGCCTAGC TGGTCTGAGA GGATGATCAG 57 301 CCACACTGGA ACTGAGACAC GGTCCAGACT CCTACGGGAG GCAGCAGTGG GGAATATTGG 361 GCAATGGGCG AAAGCCTGAC CCAGCAACGC CGCGTGGAGG AAGACACCCT TCGGGGCGTA 421 AACTCCTTTT ATAGCCGAAG TTATGACGGT AGGCTATGAA TAAGCACCGG CAAACTCCGT 481 GCCAGCAGCG CGGTAATACG GAGGGTGCAA GCGTTACTCG GAATCACTGG GCGTAAAGAG 541 CGCGCAGGCG GTATTGTAAG CTAGATGTGG AAGCCTACAG CTCAACTGTA GAACTGCATC 601 TAGAACTGCG ATACTAGAGT CTGGGAGAGG TAACTGGAAT TAGTGGTGTA GGGGTAAAAT 661 CCGTAGAGAT CACTAGGAAT ACCGAAAGCG AAGGCAAGTT ACTGGAACAG TACTGACGCT 721 GAGGCGCGAA AGCGTGGGGA GCAAACAGGA TTAGATACCC TGGTAGTCCA CGCCCTAAAC 781 GATGGATGTT AGTCGTTGGG AAGCTAGTCT TCTCAGCGAT GCAGCTAACG CATTAAACAT 841 CCCGCCTGGG GAGTACGGCC GCAAGGTTAA AACTCAAAGG AATAGACGGG GACCCGCACA 901 AGTGGTGGAG CATGTGGTTT AATTCGAAGA TACGCGAAGA ACCTTACCCA GCCTTGACAT 961 CGATGGAACC CGCTAGAAAT AGTGGGGTGC CCCTTTTGGG GAGCCTGAAG ACAGGTGCTG 1021 CACGGCTGTC GTCAGCTCGT GTCGTGAGAT GTTGGGTTAA GTCCCGCAAC GAGCGCAACC 1081 CTCATGCTTA GTTACTAACA GTTCGGCTGA GGACTCTAAG CAGACTGCCC GGGCAACCGG 1141 GAGGAAGGTG GGGACGACGT CAAGTCATCA TGGCCCTTAC GGCTGGGGCG ACACACGTGC 1201 TACAATGGGT AGGACAGAAA GATGCAATAC TGCGAAGTGG AGCAAAACTC TAAACCTACT 1261 CTCAGTTCGG ATTGTAGTCT GCAACTCGAC TACATGAAGC TGGAATCACT AGTAATCGCG 1321 AATCAGCCAT GTCGCGGTGA ATACGTTCCC GGGTCTTGTA CTCACCGCCC GTCACACCAT 1381 GGGAGTTGAG TTCACCCGAA GCGGGGATGC TAAAACAGCT ACCCTCCACG GTGGGCTCAG 1441 CGACTGGGGT GAAGTCGTAA CAAGGTAACC GTAGGAGAAC CTGTGGCTGG ATCACCTCCT 1501 TA 58 APPENDIX 4 LOCUS APG56B 1504 by DNA 11-AUG-1994 DEFINITION Epibiont of Alvinella pompejana of 16S rRNA gene encoding 16S ribosomal RNA. KEYWORDS 16S ribosomal RNA. SOURCE Epibiont of Alvinella pompejana, 5A from APG. ORGANISM Epibiont of Alvinella pompejana Bacteria /Prokaryota; Proteobacteria; subgroup: epsilon. REFERENCE 1 (bases 8 to 1542) AUTHORS Haddad,M.A., Camacho,F. and Cary,S.C. TITLE Phylogenetic characterization of the epibiotic bacteria associated with the hydrothermal vent polychaete, Alvinella pompejana JOURNAL in prep, 0-0 (1994) STANDARD full automatic COMMENT Data kindly submitted in computer readable form by: S. Craig Cary Ph.D Microbiology Oregon State University Corvallis Oregon 97331 USA Phone: (503) 737-1837 Email: caryc@ava.bcc.orst.edu Fax: (503) 737-0496 [1] Author requested hold until 01-OCT-1994 FEATURES Location/Qualifiers rRNA 1..1504 /gene="16S rRNA" /product="16S ribosomal RNA" BASE COUNT 397 a 341 c 459 g 305 t 2 others ORIGIN 1 AGAGTTTGAT CATGGCTCAG AGTGAACGCT GGCGGCGTGC TTAACACATG CAAGTCGAAC 61 GGAAACAGGC CCTTCGGGGT GCTGTCGAGT GGCGCACGGG TGAGTAATGT ATAGCTAACA 121 TGCCCTTTGG AGGGGGATAG CCACTGGAAA CGGTGATTAA TACCCCATAC TCCTCGAGAA 181 CTTAAGTTCT TGAGGGAAAT CTTTTTTGGC CAAAGGATTG GGCTATATAG TATCAGCTTG 241 TTGGTGAGGT AACGGCTCAC CAAGGCTATG ACGCTTAGCT GGTCTGAGAG GATGACCAGC 59 301 CACACTGGAA CTGAGACACG GTCCAGACTC CTACGGGAGG CAGCAGTGGG GAATATTGGG 361 CAATGGGCGG AAGCCTGACC CAGCAACGCC GCGTGGAGGA AGAAACCCTT AGGGGCGTAA 421 ACTCCTTTTA TCAGGGAAGA TAATGACGGT ACCTGATGAA TAAGCACCGG CTAACTCCGT 481 GCCAGCAGNC GCGGTAATAC GGAGGGTGCA AGCGTTACTC GGAATCACTG GGCGTAAAGA 541 GCGCGCAGGC GGTTTGGCAA GCTAGATGTG AAAGCCTGTA GCTCAACC AC AGAACTGCAT 601 CTAGAACTGC AGACTAGAGT CTGGGAGGGG TAGCTGGAAT TAGTGGTGTA GGGGTAAAAT 661 CCGTAGAGAT CACTAGGAAT ACCGAAAGCG AAGGCGAGCT ACTGGAAC AG TACTGACGCT 721 GAGGCGCGAA AGCGTGGGGA GCAAACAGGA TTAGATACCC TGGTAGTCCA CGCCCTAAAC 781 GATGGATGTT AGTCGTTGGA GAGCAAGTCT CTCCAGTGAT GCAGCTAACG CATTAAACAT 841 CCCGCCAGGG GAGTACGGTC GCAAGACTAA AACTCAAAGG AATAGACGGG GACCCGCACA 901 AGTGGTGGAG CATGTGGTTT AATTCGAAGA TACGCGAAGA ACCTTACCCA GTCTTGACAT 961 CGATGGAACC TGCTAGAGAT AGCGGGGTGC CCCTTTTGGG GAACCTGAAG ACAGGTGCTG 1021 CACGGCTGTC GTCAGCTCGT GTCGTGAGAT GTTGGGTTAA GTCCCGCAAC GAGCGCAACC 1081 CTCATCCTTA GTTGCTAACA GGTTTGGCTG AGAACTCTAA GGAGACTGCC CGGGCAACCG 1141 GGAGGAAGGT GGGGATGACG TCAAGTCATC ATGGCCCTTA CGGCTGGGGC GACACACGTG 1201 CTACAATGGG CAGGACAGAG AGAAGCAATA CCGCGAGGTG GAGCAAAGCT GTTAAACCTG 1261 CTCTCAGTTC GGATTGTAGT CTGCAACTCG ACTACATGAA GTTGGAATCA CTAGTAATCG 1321 CGAATCAGCC ATGTCGCGGT GAATACGTTC CCGGGTCTTG TACTCACCNC CCGTCACACC 1381 ATGGGAGTTG AG'TTCACCCG AAGCGGGGAT GCTAAAATAG CTACCCTCCA CGGTGGGCTC 1441 AGCGACTGGG GTGAAGTCGT AACAAGGTAA CCGTAGGAGA ACCTGTGGTT GGATCACCTC 1501 CTTA