Origin of Chemical Shifts BCMB/CHEM 8190
advertisement

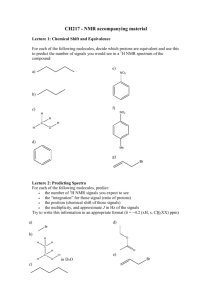
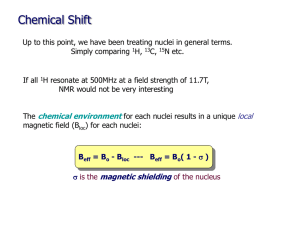
Origin of Chemical Shifts BCMB/CHEM 8190 Empirical Properties of Chemical Shift υi (Hz) = γB0 (1-σi) /2π σi, shielding constant dependent on electronic structure, is ~ 10-6. Measurements are made relative to a reference peak (TMS). Offsets given in terms of δ in parts per million, ppm, + downfield. δi = (σref - σi ) x 106 or δi = (( υi - υref )/υref) x 106 Ranges: 1H, 2H, 10 ppm; 13C, 15N, 31P, 300 ppm; 19F, 1000 ppm Ramsey’s Equation for Chemical Shift • Additional Reference: G.A. Webb, in “Nuclear Magnetic Shielding and Molecular Structure”, J.A. Tossel, ed. Nato Adv. Sci. Series (1993) 1-25 • Physical origin: moving charges experience a force perpendicular to the trajectory. Hence electrons precess. • Circulating current gives an opposing field. B0 F r e- v F = - (e/c) v x B0 B’ = -(e/c) (r x v) / r3 = -(e/cm) (r x p) / r3 • But, we actually need to treat electrons at QM level Some Quantum Mechanics Fundamentals Expectation values correspond to observables: O = <ψ| O |ψ> = ∫ ψ* O ψ dτ O - an operator, ψ - a wave function (electronic or spin) Examples, wave functions: ψ = 1s, 2s, 2p1, 2p0, 2p-1…… (electronic wave functions) ψ = α, β (one spin ½ ), αα, αβ, βα, ββ (two spins ½ ) All are solutions to Schrodinger’s equation: H ψ = E ψ They are normalized: <ψ|ψ> = ∫ ψ*ψ dτ = 1 Examples, Operators: Hamiltonian operator is special: <ψ| H |ψ> = E Zeeman Hamiltonian for nuclei in a magnetic field: Hz = - μ•B0, Ez = < α|- μ•B0 |α> Begin with classical expression: substitute QM operators μz = γ Iz (h/2π) (magnetic moment) Ez = < α|-(γh/2π)IzB0 |α> = -½ γ(h/2π) B0<α*|α> = -½ γ(h/2π)B0 Quantum Expression for B’ • Have QM operator for linear momentum: p0 = i(h/(2π))(∂/∂x + ∂/∂y +∂/∂z) • But momentum in magnetic field has a “curl” p = p0 + e(B x r) / (2c) = p0 + (e/c) A A is the vector potential; A = (B x r)/2 B’ = -(e/cm) (r x p0) / r3 - e2 (r x A) / (r4c2m) • Quantum mechanically: B’ = <ψ0| -e (r x p0)/r3(cm) - e2(r x A)/(r3c2 m) |ψ0> paramagnetic diamagnetic Diamagnetic Shifts • Note: only the second term is proportional to B0 at first order theory; this is the diamagnetic term; Lamb term B’D = <ψ0| - e2(r x B0 x r)/(2r3c2 m) |ψ0> Only interested in the z component: k (x ⋅ (B0 x r)y – y ⋅ (B0 x r)x) i j k x ( Bxr ) x y ( Bxr ) y z ( Bxr ) z B’D = - (e2/(2c2 m))B0 ∫ψ0*| (x2 + y2)/r3) |ψ0 dτ Predictions: depends on electron density near to nucleus opposes magnetic field (shields) Examples: He 2 1s eσ = 59.93 x 10-6 Ne 10 eσ = 547 x 10-6 H ~2 1s eσ = ~60 x 10-6 HO- O withdraws ~10% ~6 ppm downfield Paramagnetic Contribution to Shifts • This comes from the first term – there was no explicit B0 dependence – so carry to second order electronic wave function is changed by field • B0 can be in H’ ψ = ψ0 + Σn (<ψn| H’ |ψ0> / (En-E0)) ψn = ψ0 + ψ’ H0 = (1/(2m)) p02 + V … in absence of field H = (1/(2m)) (p0 + (e/c)A) 2 + V … in presence A = (B0 x r)/2 …. This introduces field dependence H’ = (e/(2mc)) A⋅p0 … keeping most important term Paramagnetic term continued A⋅p0 = ((B0 x r)/2)⋅p0 ≡ B0⋅(r x p0)/2 = B0⋅(Lh/(2π))/2 = B0Lzh/(2π))/2 H’ = B0eh/(8πmc) Lz Hence ψ’ ∝ Σn (<ψn| LZ |ψ0> /ΔE) B’P = <ψ0 + ψ’| (e/(cm))(r x p0)/r3 |ψ0 + ψ’> = <ψ0 + ψ’| (e/(cm))(Lz)/r3 |ψ0 + ψ’> Substituting ψ’ and saving only terms linear in B0 B’P ∝ Σn[(<ψ0|Lz|ψn><ψn|Lz/r3|ψ0>)/(En-E0) + (<ψ0|Lz/r3|ψn><ψn|Lz|ψ0>)/(En-E0)] Implications for Paramagnetic Term • σP is negative (B’P ∝ - σP) … opposite to σD • σP is zero unless LZ |ψ> is finite hence, if only “s” orbitals populated, LZ |”s”> = 0 hence, small shift range for 1H • 13C has “p” orbitals (Lz |p1> = 1 p1) and finite σP • Electron distribution must also be assymetric otherwise, Σ Lz |p> = 0 hence, CH4 shift is small and resonance far upfield 13C Example: Ethane vs Ethylene • CH3-CH3 6 ppm, CH2= CH2 123 ppm, Why? • σD is about the same for both, ~200 x 10-6 • σP ∝ Σn[(<ψ0|Lz|ψn><ψn|Lz/r3|ψ0>)/(En-E0) + .. • Examine ψn = ΣicinφI, φI = 1sc, 2sc, 2pc0, 2pc+/-1, 1sH • Only ps count, ΔE small is most important • Consider first excited state: π* = (1/√2)(piA-piB) Consider Field Parallel to C-C Bond B0 A B π* = (1/√2)(pxA-pxB) π* = (1/√2)((p1A+p-1A)-(p1B+p-1B)) • Lz| π*> = (ih√2/π)((p1A-p-1A)-(p1B-p-1B))/(2i) = (ih√2/π)(pyA-pyB)> • <ψ0|Lz| π*> is finite if pyA, pyB are populated in ψ0 • ψ0 must also be assymetric – look at MOs Molecular Orbitals for Ethylene ψ3, 3 nodes, 1 bond ψ2, 2 nodes, 4 bonds E ψ1, 1 node, 5 bonds • Fill with electrons: 2x6 for C, 4 for H = 16 • 4 in 1sC, 2 in π0 (⊥ to plane), 2 in C-C σ, 4 in C-H σ • Implies 4 maximum in above Calculating Paramagnetic Contribution • Only ψ1, ψ2, contribute • ψ1, is symmetric, implies <ψ1|Lz| π*> is zero • ψ2, is asymmetric and counts • σP = -(eh/(2πmc))2<(1/r3)>2p c22 ≅ -200 x 10-6 • σc-c = σD + σP = (200-200) x 10-6 = 0 x 10-6 What about Field Perpendicular to Plane? B0 π* = (1/√2)(pzA-pzB) π* = (1/√2)(p0A-p0B) A B • Lz| p0> = 0; therefore, σP = 0 • σ⊥ = σD + σP = (200 +0) x 10-6 • Similar for in plane, perpendicular to pπs • σ (predict) = ⎡0 ⎤ (observe) ⎡− 20 ⎢ ⎢ ⎢⎣ 0 ⎥ ⎥ 200⎥⎦ ⎢ ⎢ ⎢⎣ ⎤ ⎥ 120 ⎥ 200⎥⎦ • Isotopic shift = 1/3 Tr σ = 70-100 ppm below Me • Waugh, Griffin, Wolff, JCP, 67 2387 (1977) – solids NMR 13C Chemical Shift Calculations on Peptides α helix, -57, -47; β sheet, -139, 135; Oldfield and Dios, JACS, 116, 5307 (1994) 13C • • • • shifts and Peptide Geometry Shifts relative to random coil with same amino acid Spera and Bax, JACS, 113, 5490 (1991) See also: Case, http://www.scripps.edu/mb/case (Shifts) See also: Wishart, http://redpoll.pharmacy.ualberta.ca/shiftz Remote Group Effects B0 C O H deshielded B’ B’ H shielded benzene does the same thing • σ’remote = Δχ/r3 (1-3cos2θ) • Benzene protons are 2 ppm further downfield • Johnson and Bovey, JCP, 29, 1012 (1962) Shielding from a Benzene Ring Recent Applications of Chemical Shift to Protein Structure Determination • Shen Y, Bax A, Protein backbone chemical shifts predicted from searching a database for torsion angle and sequence homology JOURNAL OF BIOMOLECULAR NMR 38: 289302,2007 • Cavalli A, Salvatella X, Dobson CM, et al. Protein structure determination from NMR chemical shifts PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA 104: 9615-9620, 2007 • Shen Y, Lange O, Delaglio F, et al. Consistent blind protein structure generation from NMR chemical shift data, PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA 105: 4685-4690, 2008 • Han, B., Liu, Y. F., Ginzinger, S. W. & Wishart, D. S. (2011). SHIFTX2: significantly improved protein chemical shift prediction. Journal of Biomolecular NMR 50, 43-57. Other data can be combined with Chemical Shifts – Protein Targets now up to 25 kDa Comparison of traditional NMR structure and predicted structure with chemical shift and RDC data. Srivatsan, Lange, Rossi, et al. Science, 327:1014-1018, 2010