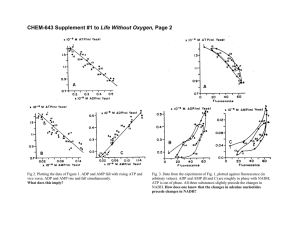
Problem 1. (2 points off for each 3 wrong)
advertisement

Problem 1. (2 points off for each 3 wrong) Problem 2 Problem 3 Complex I NADH:ubiquinone oxidoreductase net rxn: NADH + 5H+N +Q6QH2 + NAD+ + 4H+P Mass : 850 kD in 42 polypepides, FMN prosthetic groups and FeS centers Complex II Succinate dehydrogenase complex net rxn: Succinate +Q 6Fumarate + QH2 (Book usually shows as FAD and FADH2 but these are bound to the enzyme so they cannot leave. In reality the reducing equivalents are passed to ubiquinone QH2) Mass 140 kD in 5 polyptptides, FAD prosthetic group and FeS centers in proteins Complex III Cytochrome bc1 complex or ubiquinone:cytochrome c oxidoreductase net rxn: QH2 + 2Cyt c1(ox) +2H+N 6Q + 2Cyt c1(ox) + 4H+P Actually 2 reactions: QH2 + Cyt c1(ox) 6Q@- + 2H+P + Cyt c1(ox) QH2 +Q@- +2H+N + Cyt c1(ox) 6QH2 + 2H+P + Cyt c1(ox) Mass 250 kDa in 11 polypeptides, Heme and FeS groups involved Complex IV Cytochrome oxidase net rxn: 4 Cyt c1(ox) + 8H+N +O2 64 Cyt c1(ox) + 4H+P + 2H2O Mass 160 kD in 13 polypeptides, Hemes and CuA and CuB involved ATP Synthase net rxn: ADP + Pi + 3H+P 6ATP + 3H+N F1 "3$3(*, the ATP syntase FO ab2c10-12 The proton transporter proton transport down its chemiosmotic gradient is mechanically linked to changing conformations in the F1 subunit to make the enzyme first bind ADP and PI then fuse these into ATP then kick the ATP out of the active site 4. I was looking for a diagram like figure 5-11 from your text, or the following reactions Glycogen + phosphate 6 Glucose-1-P (enzyme is phosphorylase) Glucose-1-P 6Glucose-6-P (Enzyme is phosphoglucomutase) Fructose + ATP 6Frustose-6-phosphate (Enzyme is hexokinase) Galactose 6UDP-Galactose 6UDP-Glucose6Glucose-1-P6 Glucose-6-P Mannose + ATP 6Mannose-6-Phosphate (Hexokinase) Mannose-6-P 6Fructose-6-P (phosphomannose isomerse) 5. I was looking for a diagram like figure 16-6 from your text, but instead of pyruvate, insert "-ketoglutarate There are three enzymes in the complex, E1, E2, and E3 The E1 structure for pyruvate and "-ketoglutarate are structurally similar, but the sequences are different the E2 complexes are similar the E3 complexes are almost identical Overall Mech 6. The way NADH equivalents in the cytoplasm get into the mitochondria is through the Malate-Aspartate Shuttle. In the cytosol the NADH is used to reduce oxaloacetate to malate, then the malate is transported to the mitochondira where the reverse reaction occurs, and the H that started on cytosol NADH gets placed on mitochondiral NADH Then there are other steps involving glutamate, "-ketoglutarate and aspartate that are used to keep the net amount of oxaloacetate in the mitochondria from increasing. The bottom line from this shuttle system is that the reducing equivalents from the NADH are moved into the mitochondira without physically transporting the NADH into the mitochondria. Thus the 13C label that stays in the NADH molecule cannot get into the mitochondira, but the 3H labeled H atoms are transferred to Malate, and malate is transferred into the mitochondira where the tritium label now appears in the matrix.