Supportive Breeding and Inbreeding Effective Number: Reply to Nomura
advertisement

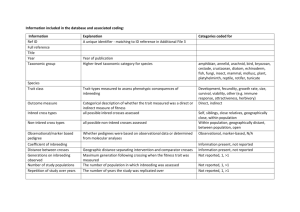
Supportive Breeding and Inbreeding Effective Number: Reply to Nomura NILS RYMAN,* PER ERIK JORDE,** AND LINDA LAIKRE Division of Population Genetics, Stockholm University, S-106 91 Stockholm, Sweden Introduction The discrepancy between Nomura’s (1998) formula for inbreeding effective number (NeI ) and our original one (Ryman & Laikre 1991) is due to different definitions of NeI. Therefore, contrary to what is implied by Nomura, neither approach is necessarily erroneous. Nevertheless, we consider ours most appropriate for practical management applications because it bears a direct relationship to the actual degree of inbreeding (and thereby to potential inbreeding depression) which the inbreeding effective number is typically thought to do. We also consider Nomura’s concept of the “total effect” of supportive breeding to be misleading because it is not clear how such a quantity should be defined. The issue may be regarded as conceptually intricate, and, because of the direct implications for management, we provide a detailed explanation of the differences between the two approaches. We introduced the concept of supportive breeding (Ryman & Laikre 1991) to address practical genetic problems encountered when weak natural populations are managed through the release of individuals bred in captivity (Fig. 1). Previously, it was largely considered genetically harmless to stock a population with individuals from the same gene pool, but our results showed that supportive breeding may result in a significant increase of inbreeding in the total (wild 1 captive) population which would not occur had it been left alone. We provided a formula for assessing the inbreeding effective population size (NeI ) of a nonselfing population subject to supportive breeding, and in a subsequent paper (Ryman et al. 1995) we presented an expression for variance effective number (NeV) and compared the immediate effect of supportive breeding on inbreeding and genetic drift. A primary intention of these studies was to provide an analytical * email nils.ryman@popgen.su.se ** Current address: Division of Zoology, Department of Biology, University of Oslo, P.O. Box 1050 Blindern, N-0316 Oslo, Norway. Paper submitted August 24, 1998; revised manuscript accepted December 28, 1998. background for supplying management recommendations on how to avoid a sudden excessive increase of inbreeding and drift in particular generations. Nomura (1998) proposes an alternate formula for inbreeding effective number which is identical to our expression for NeV, a result indicating that the impact of supportive breeding is the same on both effective numbers. He suggests that our comparisons of the effect of supportive breeding on NeI and NeV may be misleading because our formula for NeI does not account for the “total effect” of supportive breeding, whereas the one for NeV does. Clearly, Nomura’s derivations demonstrate that, using his definition of NeI , the overall effect of supportive breeding can be considered the same on NeI and NeV . This result is expected because Nomura’s definition disregards inbreeding that is due to the nonrandom mating caused by population subdivision in generation t 1 1. Under random mating, inbreeding and drift are both due to the same process of random sampling of genes during reproduction, and when the appropriate generations are considered the corresponding effective sizes should coincide. Reduction of heterozygosity and inbreeding then follows directly from the change of gene frequency caused by genetic drift. Inbreeding and drift may become manifest in different generations, and NeI may differ from NeV when population size is changing. Nevertheless, in the long run the two quantities are expected to be the same on average when mating is random. Definitions of Inbreeding Effective Size Nomura’s definition of NeI differs from ours in that it does not relate directly to the change of inbreeding that actually occurs from one particular generation to the next. When deriving our formula we applied the common definition of NeI : the inbreeding effective size of a population is defined as the size of an ideal population exhibiting the same rate of inbreeding (F ) as the actual one (Crow & Kimura 1970). Letting nF denote the 673 Conservation Biology, Pages 673–676 Volume 13, No. 3, June 1999 674 Supportive Breeding and Inbreeding Effective Number Ryman et al. Figure 1. Schematic representation of a supportive breeding situation. Before breeding, the total population in generation t 1 1 is divided into a captive and a wild group of size Nc,t 1 1 and Nw,t 1 1 that reproduce in captivity and in the wild, respectively. The Nc,t 1 2 and Nw,t 1 2 offspring are mixed before breeding and mate at random in generation t 1 2. Nt 1 1 5 Nc,t 1 1 1 Nw,t 1 1 and Nt 1 2 5 Nc,t 1 2 1 Nw,t 1 2 . Modified from Ryman et al. (1995). change of average inbreeding between two consecutive generations (t 2 1 to t), Ft – Ft – 1 ∆F t = ---------------------, 1 – Ft – 1 (1) and temporarily dropping the t subscript the inbreeding effective number is defined as 1 N eI = ----------. 2∆F (2) In contrast, Nomura uses a definition based on the change of average coancestry (nu), such that 1 N eI ,θ = ----------, 2∆θ (3) the u subscript added to mark the distinction from the NeI defined in equation 2. Citing Caballero and Hill (1992), Nomura states that in nonrandom mating populations “the rate of inbreeding Conservation Biology Volume 13, No. 3, June 1999 should be computed with the average coancestry among individuals instead of the actual inbreeding coefficient.” Although such an approach may be appropriate in some instances, we have difficulty seeing that it represents a universal truth. When interest is focused on the actual change of inbreeding (nF ) or on inbreeding depression, it seems more natural to use the traditional definition of NeI (equation 2). It is interesting to note in this context that Wright (1922) in his original treatment of inbreeding considered it important that the inbreeding coefficient relate directly to contemporary inbreeding depression. In a subdivided population, inbreeding increases for two different reasons, because of the finite size of the total population (i.e., drift) and because breeding is restricted to within subpopulations (i.e., nonrandom mating). In the absence of immigration, the inbreeding due to finite population size is “permanent,” whereas the inbreeding caused by subdivision may be considered “temporary” because it disappears if subdivision is removed and random mating established. In some instances it may be helpful to distinguish between permanent and temporary inbreeding. For such situations Caballero and Hill (1992) have shown that the inbreeding effective size defined on the basis of coancestry (NeI,u, equation 3) bears a direct relationship to the inbreeding due to the finite size of the total population. Thus, although NeI is connected with the change of total inbreeding, NeI,u relates only to the portion explained by restricted population size, ignoring the part due to nonrandom mating. To illustrate the difference between NeI and NeI,u numerically, we have simulated the accumulation of inbreeding for the Pacific salmon supportive breeding situation used as an example by Nomura (1998), keeping the same designation of generations as in his paper (Table 1). In generation t 1 1, a population of 50 breeders is divided into a captive and a wild group of Nc,t 1 1 5 30 and Nw,t 1 1 5 20 breeders, and these two groups produce Nc,t 1 2 5 5000 captive and Nw,t 1 2 5 50 wild progeny, respectively. In t 1 2 the Nt 1 2 5 5050 breeders mate at random and produce a total of Nt 1 3 5 50 progeny, and this population size of 50 panmictic breeders is maintained in subsequent generations. Our primary interest refers to nonselfing species (such as the Pacific salmon), but for comparisons with exactly the situation modeled by Nomura we also conducted simulations permitting a random amount of selfing. When selfing is excluded there is the usual delay of one generation before inbreeding is manifest, because individuals cannot be homozygous for alleles that are identical by descent from generation t 1 1 until generation t 1 3 (Table 1, Fig. 1). The change of total inbreeding experienced in t 1 3 (nFt 1 3) corresponds exactly to that predicted by our original formula for NeI (Ryman & Laikre 1991), such that nFt 1 3 5 1/(2NeI, t 1 1), as it should (equation 2). Similarly, the inbreeding coefficient Ryman et al. Supportive Breeding and Inbreeding Effective Number 675 Table 1. Results of pseudo-random number computer simulations of the accumulation of inbreeding (F) in selfing and nonselfing populations subject to supportive breeding.a No selfing Generation t11 t12 t13 t14 Nc Nw F 30 20 5000 50 5050d 50 50 0 DF Selfing NeI 5 1/(2DF) 30.6c 0 0 0.01634 0.01644 0.01634 0.00010 5134.3 F DF 0 NeI 5 1/(2DF) 29.9 0.01675 0.01675 0.01644 0.02627 20.00032 0.01000 21572.8 50 Nomura’s NeIb 50 76.9 50 a Cf. Fig. 1 and Nomura (1998). All individuals are uniquely heterozygous in generation t 1 1 ( Ft11 5 0), and subsequent generations were created through random sampling of the number of gametes necessary to create the stipulated number of diploid offspring. Simulated F values represent the mean of one million replicates, and deviations between those means and the ones calculated exactly are noticeable in the sixth decimal place only. The NeI estimates for particular generations are based on the D F observed one and two generations later in the case of selfing and nonselfing, respectively. b From Table 1 in Nomura (1998); the value designed NeI, t11→t12 is assigned to generation t 1 1, etc. c The formula of Ryman and Laikre (1991) yields 30.60. d Captive and wild progeny mix and mate at random in t 1 2. In subsequent generations all matings take place in the wild and there is no further release of captive progeny. in t 1 4 is determined both by the number of breeders in t 1 2 and by the inbreeding in t 1 3 through 2N t + 2 – 1 1 F t + 4 = ---------------- + ------------------------- F t + 3 2N t + 2 2N t + 2 (4) (Crow & Kimura 1970; equation 3.11.3). Thus, in accordance with expectation for the early generations, the increase of F from t 1 3 to t 1 4 corresponds approximately, but not exactly, to the number of breeders in t 1 2 [1/(2nFt 1 4) 5 5134.3 rather than 5050; Table 1]. In the case where a random amount of selfing occurs, the split in t 1 1 has the effect that substantial inbreeding due to supportive breeding is manifest already in t 1 2. In some situations, such as the present one, there is even a reduction of inbreeding from t 1 2 to t 1 3. The pattern for accumulation of inbreeding is quite different when selfing is allowed or disallowed in the early generations following an event of supportive breeding. Most important, however, Nomura’s NeI,u of 76.9 that is based on coancestry does not correspond to the change of total inbreeding (nF) between any two generations, regardless of selfing or nonselfing (Table 1). Rather, 76.9 corresponds to the change of coancestry (nu ) from t 1 1 to t 1 2 where ut 1 1 5 1/(2Nt 1 1) and ut 1 2 5 Ft 1 3. It should be noted that using these definitions ut 1 1 is different from the actual level of inbreeding in t 1 2 (Ft 1 2 5 0.01675): the inbreeding due to nonrandom mating in t 1 1 is disregarded. “Total Effect” of Supportive Breeding We consider the issue of defining a total effect of supportive breeding potentially less straightforward than indicated by Nomura. An obvious problem encountered when trying to compare “total” effects on drift and actual inbreeding (F) is that they become manifest in dif- ferent generations. For a nonselfing organism the effect on drift is displayed in generation t 1 2 (Fig. 1), whereas an effect on inbreeding does not occur until t 1 3. Our formula for NeI (Ryman & Laikre 1991) relates to the increase of inbreeding that is manifest in t 1 3, and this change (nFt 1 3 ) depends only on the number of wild and captive parents in t 1 1 and their relative contribution to t 1 2. Nomura argues that the number of individuals in t 1 2 (Nt 1 2 ) is also an effect of supportive breeding that should be accounted for when evaluating the total effect. We have not “neglected” this aspect, but it is not clear to us why the number in t 1 2 is the only one that should be considered. The goal of a supportive breeding operation typically is to increase the population size over several generations, and we see no reason why the total effect should account for only two generations as a general rule. The example chosen by Nomura (Table 1) depicts only one of a series of possible scenarios: a population that increases dramatically in t 1 2 and then crashes to its original size in t 1 3. In this particular case it may seem natural to consider the larger size in t 1 2 and the correspondingly smaller nFt 1 4. In other situations, however, there may be no crash or the crash may occur much later. It would then seem equally natural to regard the larger size persisting over multiple generations as an effect of successful supportive breeding. The rate of inbreeding would then be reduced over a range of subsequent generations, and it is not obvious how to define a generation in which the effect of supportive breeding ends. In other situations supportive breeding may be at least partially successful yet not result in an increased number of breeders in t 1 2. This would occur, for example, in a salmonid population in which the area available for spawning can accommodate only a restricted number of redds. Releasing large numbers of hatchery juveniles might then result in a disproportionately large fraction of captive progeny in the pre-spawning population, and Conservation Biology Volume 13, No. 3, June 1999 676 Supportive Breeding and Inbreeding Effective Number this over-representation may persist among the breeders. In such a case supportive breeding might be considered successful because of the larger abundance of fish, although the absolute number of breeders remains the same as in previous generations. Of course, a manager who is interested in the average generational change of inbreeding from t 1 2 to t 1 4 may compute that quantity (and the average of nFt 1 3 and nFt 1 4). Here, nFt 1 3 follows from the formula of Ryman and Laikre (1991), and nFt 1 4 is given by equation 4. For the simulation in Table 1 the average nF is (0.01634 1 0.00010)/2 5 0.00822, corresponding to an average NeI of 60.83; this approximate number can also be obtained through the harmonic mean of the corresponding NeI s (the harmonic mean of 30.6 and 5134.3 is 60.84; Crow & Kimura 1970). We see a potential danger, however, in dealing with an NeI that represents an average over multiple generations rather than with the separate ones reflecting the change from one generation to another. First, in followup studies of a supportive breeding project it may be imperative to have accurate information about the actual inbreeding of each particular generation. For the operation pictured in Table 1, for example, the inbreeding “strikes” in t 1 3, whereas the change from t 1 3 to t 1 4 is negligible. Thus, if the actual values of nF are known, the observation of, for example, a high mortality in t 1 3 may correctly alert a manager that the population may be notably susceptible to inbreeding depression. In contrast, the possibility of inbreeding depression may go unnoticed if inbreeding is erroneously believed to increase uniformly over both generations. Further, supportive breeding projects should be planned to avoid an excessive increase of inbreeding in any particular generation. For example, in line with general recommendations for conservation genetics (Frankel & Soulé 1981), we suggested that operations resulting in an NeI Conservation Biology Volume 13, No. 3, June 1999 Ryman et al. smaller than 50 should be modified to provide less dramatic increases in inbreeding (Ryman et al. 1995). In this respect the Pacific salmon scenario (Table 1) must be considered a failure in that the inbreeding being expressed in t 1 3 corresponds to an NeI of only about 30. By using our NeI formula when planning the operation, such a large increase of inbreeding could have been avoided by modifying the number of wild and captive breeders in t 1 1 and their proportionate contribution to t 1 2. In contrast, Nomura’s equation does not provide this opportunity. Acknowledgments This note was prepared with support from the Swedish Natural Science Research Council, and part of the work was conducted within the scope of the Swedish research program on Sustainable Coastal Zone Management, SUCOZOMA, funded by the Foundation for Strategic Environmental Research, MISTRA. Literature Cited Caballero, A., and W. G. Hill. 1992. Effective size of nonrandom mating populations. Genetics 130:909–916. Crow, J. F., and M. Kimura. 1970. An introduction to population genetics theory. Harper & Row, New York. Frankel, O. H., and M. E. Soulé. 1981. Conservation and evolution. Cambridge University Press, Cambridge, United Kingdom. Nomura, T. 1998. Effective population size in supportive breeding. Conservation Biology, 13:670–672. Ryman, N., and L. Laikre. 1991. Effects of supportive breeding on the genetically effective population size. Conservation Biology 5:325–329. Ryman, N, P. E. Jorde, and L. Laikre. 1995. Supportive breeding and variance effective population size. Conservation Biology 9:1619–1628. Wright, S. 1922. Coefficients of inbreeding and relationship. American Naturalist 56:330–338.