The OLS estimator (b.l.u.e.) for is = ( )
advertisement

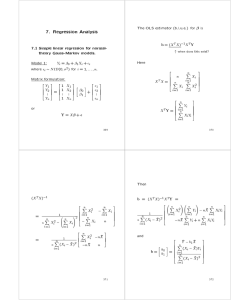
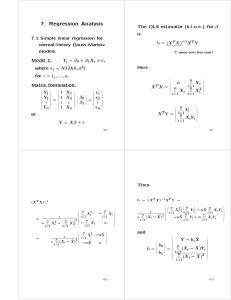
The OLS estimator (b.l.u.e.) for is
7. Regression Analysis
b = (X T X );1X T Y
7.1 Simple linear regression for normal-theory
Gauss-Markov models.
" when does this exist?
Here
Model 1:
Yi = 0 + 1Xi + i
where 1 NID(0; 2) for i = 1; : : : ; n.
2
66 n
T
X X = 666 X
n
4 Xi
Matrix formulation:
2Y 3 21 X 3
2 3
66 Y12 77 66 1 X12 77 " 0 # 66 12 77
64 .. 75 = 64 .. .. 75 + 64 .. 75
1
Yn
1 Xn
n
or
i=1
3
Xi 77
77
i=1
n
X
X 2 75
n
X
i=1
i
2X
3
n
66 Yi 77
77
=1
X T Y = 666 iX
n
4 XiYi 75
Y = X + i=1
373
374
Then
(X T X );1
b = (X T X );1X T Y
2X
3
n
n
X
66 Xi2 ; Xi 77
i=1 77
10
= n
12 666 i=1X
n
7
n
X 2 @ X A 4 ; Xi
n 5
n
Xi ;
Xi
i=1
=
i=1
i=1
2X
n
66 Xi2 ;nX
1
n
4 i=1
X
n (Xi ; X )2 ;nX
n
3
77
5
i=1
375
=
2 0 n 10 n 1
3
n
X
X
X
2
66 @ Xi A @ YiA ; nX XiYi 77
66 i=1
77
1
n
ni=1
n i=1
X
6
75
X
X
2
n (Xi ; X ) 4
;nX Yi + n XiYi
i=1
and
i=1
2
" # 666
b
b = 0 = 666
b1
64
Y ; b1X
n
X
(Xi ; X )Yi
i=1
n
X
(Xi ; X )2
i=1
i=1
3
77
77
77
75
376
Analysis of Variance:
Covariance matrix:
V ar(b) = V ar (X T X );1X T Y
n
X
i=1
(X T X );1X T (2I )X (X T X );1
=
= 2(X T X );1
3
2
;X 2
1 + X 2 2
= 2 664 n (;XX ;X ) (X 1;X ) 775
(X ;X )2 (X ;X )2
i
i
Yi2 =
=
=
YT Y
YT (I ; PX + PX ; P1 + P1)Y
YT (I ; PX )Y + YT (PX ; P1)Y + YT P1Y
%
%
SSE
i
i
Estimate the covariance matrix for b as
Sb = MSE (XTX);1
where
MSE = SSE =(n ; 2) = n ;1 2 Y(I ; PX)Y:
"
"Corrected
model"
sum of
squares
"
call this
R(1j0)
Correction
for the
"mean"
"
call this
R(0)
(i) By Cochran's Theorem, these three sums
of squares are independent chi-squared random variables.
(ii) By result 4.7, 12 SSE 2(n;2) if the model
is correctly specied.
378
377
Correction for the overall mean:
Reduction in residual sum of squares:
R(k+1; : : : ; k+q j 0; 1; : : : ; k )
= YT (I ; PX 1 )Y ; YT (I ; PX )Y
"
"
sum of squared
sum of squared
residuals for the
residuals for the
smaller model
larger model
Here
X = [ X1 j X2 ]
%
columns
columns
corresponding
corresponding
to 0; 1; : : : ; k to k+1 k+g
379
R ( 0 ) =
=
=
=
YT P1Y
YT (I ; I + P1)Y
YT I Y ; YT (I ; P1)Y
n
X
i=1
(Yi ; 0)2 ;
n
X
(Yi ; Y )2
i=1
%
sum of squared
residuals from tting
the model
Yi = + i.
The OLS estimator for
= E (Yi) is
^ = (1T 1);11T Y
= P1Y = Y
380
Reduction in the residual sum of squares for
regression on Xi:
Note that
R(0) =
=
Y T P1 Y
R(1j0) =
=
=
YT 1(1T 1);11T Y
0n 1
0n 1
X
X
;
1
= @ YiA (n) @ YiA
=
i=1
nY:2
i=1
YT (PX ; P1)Y
YT (;I + PX + I ; P1)Y
YT (I ; P1)Y ; YT (I ; PX )Y
%
sum of squared
residuals for
tting the model
Yi = + i
"
sum of squared
residuals for
tting the model
Yi = 0 + 1Xi + i
381
382
Consider the following F-tests:
ANOVA table:
Source of
variation
d.f.
Regression
on X
1
Residuals n ; 2
Corrected
total
n;1
Correction
for mean
1
Sum of
Squares
Mean
Square F
YT (PX ; P1)Y
YT (I ; PX )Y
YT (I ; P1)Y
YT P1Y = nY 2
From result 4.7 we have
1
1 T
2 2
2 R(0) = 2 Y P1Y 1( )
where
2 = 12 T XT P1X
= 12 T XT P1T P1X = 12 (P1X )T (P1X )
= n2 (0 + B1X )2
Hypothesis test: Reject H0 : 0 + 1X = 0 if
(0) > F
F = RMSE
(1;n;2); 383
384
Here
(PX ; P1)X = (PX ; P1)[1jX]
Test the null hypothesis H0 : 1 = 0
=
1j0)=1
F = R(MSE
T
2] ]
= [Y[TY(I(;PXP ;)PY1])=Y[(]n=[1
; 2)2
X
=
=
F(1;n;2)(2)
where
2 = 12 T X T (PX ; P1)X = 12 T X T (PX ; P1)T (PX ; P1)X %
The null hypothesis is
H0 : (PX ; P1)X = 0
=
2
3
4(PX ; P1)1 (PX ; P1)X5
2
3
4PX 1 ; P11 PX X ; P1X5
1
1 ; 1 X ; X
2 0 j X ; X 3
66 0 j X12 ; X 77
64 .. j
.. 75
0 j Xn ; X
If any Xi 6= Xj , then (PX ; P1)X = 0 if and
only if 1 = 0. Hence, the null hypothesis is
H0 : 1 = 0. Note that
n
X
2 = 12 12 (Xi ; X )2
i=1
385
386
Reparameterize the model:
Yi = + 1(Xi ; X ) + i
with i NID(0; 2); i = 1; : : : ; n.
Interpretation of parameters:
= E (Y ) when X = X
Maximize the power of the F-test for
H0 : 1 = 0 vs. HA : 1 6= 0
by maximizing
n
X
2 = 12 12 (Xi ; X )2
i=1
1 is the change in E (Y ) when
X is increased by one unit.
387
388
Matrix formulation:
2 3 2
X
64 Y..1 75 = 64 1.. X1 ;
.
Yn
1 Xn ; X
or
Clearly,
3" # 2 3
75 + 64 ..1 75
1
n
For this reparameterization, the columns of W
are orthogonal and
2n
3
0
77
n
W T W = 664 0 X
(Xi ; X )2 5
21
(W T W );1 = 4 0n
Y = W + "
#
W = X 01 ;1X = XF
WTY
"
#
X = W 10 ;1X = WG
i=1
0
1
(X ;X )2
3
5
i
2X
3
n
66 Yi
77
6
77
i
=1
= 66 X
n
4 (Xi ; X )Yi 75
i=1
390
389
Then,
"
^ = ^^
1
#
Analysis of variance:
= (W T X );1W T Y
The reparamterization does not change the
ANOVA table.
2
3
Y
6
= 4 (X ;X )Y 75
(X ;X )2
i
i
i
and
V ar(^ ) = 2(W T W );1
2 2
3
0
n
75
= 64
0 (X;2 X )2
i
X ;X )Y
Hence, Y and ^1 = (
(X ;X )2 are uncorrelated
(independent for the normal theory GaussMarkov model).
i
i
i
391
Note that
PX = X (X T X );1X T
= W (W T W );1W T = PW
and
R(0) + R(1j0) + SSE
= YT P1Y + YT (PX ; P1)Y + YT (I ; PX )Y
= YT P1Y + YT (PW ; P1)Y + YT (I ; PW )Y
= R() + R(1j)+ SSE
392
7.2 Multiple regression analysis for the normaltheory Gauss-Markov model
Yi = 0 + 1X1i + + r Xri + i
where
i NID(0; 2) for i = 1; : : : ; n :
Matrix formulation:
Y = X + where
" "
1
X1
(iii) Y^ = X b = X (X T X );1X T Y = PX Y
(v) By result 4.7,
1 T
1
2 SSE = 2 e e
= 12 YT (I ; PX )Y
2(n;r;1)
3
Xr1 7 2 0 3
. Xr2 77 66 1 77
77 6 . 7
..
75 4 . 5
.
Xrn r
"
"
X2
(i) the OLS estimator (b.l.u.e.) for is
b = (X T X );1X T Y
(ii) V ar(b) = 2(X T X );1
(iv) e = Y ; Y^ = (I ; PX )Y
N (0; 2I )
2
X11 X21
66 11 X
12 X.22
X = 666 1 ..
..
64 .. ..
.
1 X1n X2n
Suppose rank(X ) = r + 1, then
Xr
(vi) MSE = nSSE
;r;1 is an unbiased estimator of
2.
394
393
Reduction in the residual sum of squares obtained by regression on X1; X2; : : : ; Xr is denoted as
R(1; 2; : : : ; r 0)
ANOVA
Source of
variation
Model (regression
on X1; : : : ; Xr)
Error (or
residuals)
Corrected
total
Correction for
the mean
d.f.
r
Sum of
squares
R(1; : : : ; r j0)
= YT (PX ; P1)Y
n;r;1
YT (I ; PX )Y
n;1
1
YT (I ; P1)Y
R(0) = YT P1Y
=nY 2
= YT (I ; P1)Y ; YT (I ; PX )Y
= Y T (P X ; P 1 ) Y
Use Cochran's theorem or results 4.7 and 4.8
to show that SSE is distributed independently
of
and
1
2
2 SSE (n;r;1)
and
395
R(1; 2; : : : ; rj0) = SSmodel
1 R( ; : : : ; B j ) 2 (2)
r 0
(r)
2 1
396
Note that
Then
: : ; r j0)=r F
2
F = R(1; :MSE
(r;n;r;1)( )
where
2 = 212 T XT (PX ; P1)X = 212 T X T (PX ; I + I ; P1)X h
i
= 212 T X T (I ; P1)X ; T XT (I ; PX )X %
This is a matrix of zeros
= 212 T X T (I ; P1)X = 212 T X T (I ; P1)(I ; P1)X = 212 [(I ; P1)X ]T (I ; P1)X 2
3
4
(I ; P1)X = (I ; P1)1(I ; P1)X1 (I ; P1)Xr 5
2 3
= 4 0 X1 ; X11 Xr ; Xr15
=)
(I ; P1)X =
=)
2
r
X
j =1
j (Xj ; Xj 1)
r
X
2 = 12 4 j2(Xj ; Xj 1)T (Xj ; Xj 1)
j =1
XX
+
j k (Xj ; Xj 1)T (Xk ; Xk 1)
j 6=k
2
3
n
X
= 212 T 4 (Xj ; X )(Xi ; X )T 5 i=1
where2 3
2 3
2
3
X
X
1
1
1
i
= 64 .. 75 Xi = 64 .. 75 i = 1; : : : ; n
= 64 .. 75 X
r
Xr
Xri
398
397
n
X
If (Xi ; X )(Xj ; X )T is positive denite,
i=1
then the null hypothesis corresponding to 2 =
0 is
H0 : = 0( or 1 = 2 = = r = 0)
Sequential sums of squares (Type I sums of
squares in PROC GLM or PROC REG in SAS).
Let
Reject
H0 : = 0 if
T
; P1)Y=r > F
F = YT (IY;(PPX)Y
=(n ; r ; 1) (r;n;r;1); X
399
X0 = 1
X1 = [1jX1]
X2 = [1jX1jX2]
..
Xr = [1jX1j jXr ]
P0 = X0(X0T X0);1X0T
P1 = X1(X1T X1);X1T
P2 = X2(X2T X2);X2T
..
Pr = Xr (XrT Xr );XrT
400
YT Y
=
YT P0Y + YT (P1 ; P0)Y + YT (P2 ; P1)Y
+ + YT (Pr ; Pr;1)Y + YT (I ; Pr)Y
= R(0) + R(1j0) + R(2j0; 1)
+ + R(rj0; 1; : : : ; r;1)
+SSE
Use Cochran's theorem to show
{ these sums of squares are distributed
independently of each other.
{ Each 12 R(ij0; : : : ; i;1) has a chisquared distribution with one degree of
freedom.
Use Result 4.7 to show 12 SSE 2(n;r;1).
Then
: : : ; j ;1)=1 F
F = R(j j0;MSE
1;n;r;1(2)
where
2 = 12 T X T (Pj ; Pj ;1)X = 12 T X T (Pj ; Pj;1)T (Pj ; Pj;1)X = 12 [(Pj ; Pj;1)X ]T (Pj ; Pj;1)X Hence, this is a test of
H0 : (Pj ;Pj ;1)X = 0 vs Ha : (Pj ;Pj ;1)X 6= 0
401
Note that
(Pj ; Pj;1)X
2 = (Pj ; Pj;1)4X1 Xj ;1 3
Xj Xr 5
2
= 4(Pj ; Pj;1)X1 (Pj ; Pj;1)Xj;1 3
(Pj ; Pj;1)Xj 5
2
4
= Onj (Pj ; Pj;1)Xj 3
(Pj ; Pj ;1)Xr 5
403
402
Then
(Pj ; Pj;1)X =
r
X
k=j
k (Pj ; Pj ;1)Xk
= j (Pj ; Pj;1)Xj
+
r
X
k=j +1
and the null hypothesis is
H0 : 0 = j (Pj ;Pj ;1)Xj +
k (Pj ; Pj ;1)Xk
r
X
k=j +1
k (Pj ;Pj ;1)Xk
404
From the previous discussion:
Type II sums of squares in SAS (there are also
Type III and Type IV sums of squares for
regression problems).
R(j j0 and all otherk0 s) = YT (PX ; P;j )Y
where
P;j = X;j (X;T j X;j );X;T j
and X;j is obtained by deleting the j -th
column of X .
T
; P;j )Y=1 F
2
F = Y (PXMSE
(1;n;r;1)( )
where
2 = 12
= 12
T X T (PX ; P;j )X j2XTj (PX ; P;j )Xj
This F-test provides a test of
H 0 : j = 0
vs
HA : j 6= 0
if (PX ; P;j )Xj 6= 0.
405
406
When X1; X2; : : : ; Xr are all uncorrelated, then
Variable
Type I Sums
of squares
R(1j0)
=YT (P1 ; P0)Y
X2
R(2j0; 1)
= YT (P2 ; P1)Y
..
..
Xr
R(r j0; 1; : : : ; r;1)
= YT (Pr ; Pr;1)Y
Residuals SSE = YT (I ; PX )Y
X1
Corrected
Total
Type II Sums
of squares
R(1j other 0s)
=YT (PX ; P;1)Y
R(2j other 's)
= YT (PX ; P;2)Y
..
R(r j0; : : : ; r;1)
= YT (PX ; P;r )Y
(i) R(j j 0 and any other subset of 's)
= R(j j0)
and there is only one ANOVA table.
(ii) R(j j0) = ^j2
n
X
(Xji ; Xj:)2
i=1
j0) F
(iii) F = R(MSE
1;n;k;1(2)
n
X
where 2 = 12 j2 (Xji ; Xj:)2
i=1
and this F-statistic provides a test of
H0 : j = 0 versus HA : j 6= 0.
j
YT (I ; P1)Y
407
408
Condence interval for an estimable
function cT For any testable hypothesis, reject H0 : C = d
in favor of the general alternative HA : C 6= d
if
cT b t(n;rank(X ))=2
T
T );C T ];1(C b ; d)=m
F = (C b ;YdT)(I[;C (PX )X
Y=(n ; rank(X ))
q
MSE cT (X T X );c
Use cT = (0 0 .. 0 1 0 .. 0)
"
j -th position
to construct a condence interval
for j;1
Use cT = (1; x1; x2; : : : ; xr) to
construct a condence interval for
E (YjX1 = x1; : : : ; Xr ; xr )
= 0 + 1x1 + + rxr
X
> F(m;n;rank(X ));
where
m = number of rows in C
= rank(C )
and
b = (X T X );X T Y
410
409
/*
Prediction intervals:
A SAS program to perform a regression
analysis of the effects of the
Predict a future observation at
X1 = x1; : : : ; Xr = xr
i.e., predict
Y = 0 + 1x1 + + rxr + %
"
estimate the
estimate
conditional
this with
mean as
its mean
b0 + b1x1 + + br xr E () = 0
A (1 ; ) 100% prediction interval is
q
(cT b +0) t(n;rank(X ));=2 MSE [1 + cT(XTX);c]
where
cT = (1 x1 xr )
411
composition of Portland cement on the
amount of heat given off as the cement
hardens.
*/
data set1;
input run x1 x2 x3 x4 y;
/* label y = evolved heat (calories)
x1 = tricalcium aluminate
x2 = tricalcium silicate
x3 = tetracalcium aluminate ferrate
x4 = dicalcium silicate;
*/
cards;
1
7 26
6 60
78.5
2
1 29 15 52
74.3
3 11 56
8 20 104.3
4 11 31
8 47
87.6
5
6 33
95.9
7 52
6 11 55
9 22 109.2
7
3 71 17
8
1 31 22 44
6 102.7
72.5
412
/*
Regress y on all four explanatory
variables and check residual plots
and collinearity diagnostics */
9
2 54 18 22
10 21 47
11
1 40 23 34
12 11 66
13 10 68
93.1
proc reg data=set1 corr;
4 26 115.9
model y = x1 x2 x3 x4 / p r ss1 ss2
83.8
covb
9 12 113.2
collin;
output out=set2 residual=r
8 12 109.4
predicted=yhat;
run;
run;
proc print data=set1 uniform split='*';
var y x1 x2 x3 x4;
/*
label y = 'Evolved*heat*(calories)'
Examine smaller regression models
x1 = 'Percent*tricalcium*aluminate'
corresponding to subsets of the
x2 = 'Percent*tricalcium*silicate'
explanatory variables
*/
x3 = 'Percent*tetracalcium*aluminate*ferrate'
x4 = 'Percent*dicalcium*silicate';
proc reg data=set1;
run;
model
y = x1 x2 x3 x4 /
selection=rsquare cp aic
sbc mse stop=4 best=6;
run;
414
413
/*
Obs
Regress y on two of explanatory
variables and check residual plots
1
2
3
4
5
6
7
8
9
10
11
12
13
and collinearity diagnostics */
proc reg data=set1 corr;
model y = x1 x2 / p r ss1 ss2
covb
collin;
output out=set2 residual=r
predicted=yhat;
run;
/*
Evolved
Percent
heat
tricalcium
(calories) aluminate
78.5
74.3
104.3
87.6
95.9
109.2
102.7
72.5
93.1
115.9
83.8
113.2
109.4
7
1
11
11
7
11
3
1
2
21
1
11
10
Percent
Percent tetracalcium Percent
tricalcium aluminate dicalcium
silicate
ferrate
silicate
26
29
56
31
52
55
71
31
54
47
40
66
68
6
15
8
8
6
9
17
22
18
4
23
9
8
60
52
20
47
33
22
6
44
22
26
34
12
12
Use the GLM procedure to identify
all estimable functions
Correlation
*/
Variable
proc glm data=set1;
model y = x1 x2 x3 x4 / ss1 ss2 e1 e2 e p;
run;
415
x1
x2
x3
x4
y
x1
x2
x3
x4
y
1.0000
0.2286
-0.8241
-0.2454
0.7309
0.2286
1.0000
-0.1392
-0.9730
0.8162
-0.8241
-0.1392
1.0000
0.0295
-0.5348
-0.2454
-0.9730
0.0295
1.0000
-0.8212
0.7309
0.8162
-0.5348
-0.8212
1.0000
416
Parameter Estimates
The REG Procedure
Model: MODEL1
Dependent Variable: y
Variable
Analysis of Variance
Source
DF
Sum of
Squares
Mean
Square
Model
Error
Corrected Total
4
8
12
2664.52051
47.67641
2712.19692
666.13013
5.95955
Root MSE
Dependent Mean
Coeff Var
2.44122
95.41538
2.55852
R-Square
Adj R-Sq
0.9824
0.9736
DF
Parameter
Estimate
Standard
Error
t Value
1
1
1
1
1
63.16602
1.54305
0.50200
0.09419
-0.15152
69.93378
0.74331
0.72237
0.75323
0.70766
0.90
2.08
0.69
0.13
-0.21
Intercept
x1
x2
x3
x4
Number
Eigenvalue
Condition
Index
1
2
3
4
5
4.11970
0.55389
0.28870
0.03764
0.00006614
1.00000
2.72721
3.77753
10.46207
249.57825
DF
Type I SS
Type II SS
Intercept
x1
x2
x3
x4
1
1
1
1
1
118353
1448,75413
1205.70283
9.79033
0.27323
4.86191
25.68225
2.87801
0.09319
0.27323
418
Obs
Dep Var
y
Predicted
Value
Std Error
Mean Predict
Student
Residual
1
2
3
4
5
6
7
8
9
10
11
12
13
78.5000
74.3000
104.3000
87.6000
95.9000
109.2000
102.7000
72.5000
93.1000
115.9000
83.8000
113.2000
109.4000
78.4929
72.8005
105.9744
89.3333
95.6360
105.2635
104.1289
75.6760
91.7218
115.6010
81.8034
112.3007
111.6675
1.8109
1.4092
1.8543
1.3265
1.4598
0.8602
1.4791
1.5604
1.3244
2.0431
1.5924
1.2519
1.3454
0.00432
0.752
-1.054
-0.846
0.135
1.723
-0.736
-1.692
0.672
0.224
1.079
0.429
-1.113
Obs
Collinearity Diagnostics
---------------Proportion of Variation---------------Intercept
x1
x2
x3
x4
1
2
3
4
5
0.000005
8.812E-8
3.060E-7
0.000127
0.99987
0.00037
0.01004
0.000581
0.05745
0.93157
0.00002
0.00001
0.00032
0.00278
0.99687
0.00021
0.00266
0.00159
0.04569
0.94985
0.00036
0.00010
0.00168
0.00088
0.99730
419
0.3928
0.0716
0.5068
0.9036
0.8358
Variable
417
Collinearity Diagnostics
Pr > |t|
1
2
3
4
5
6
7
8
9
10
11
12
13
Cook's
D
-2-1 0 1 2
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|*
**|
*|
|
|***
*|
***|
|*
|
|**
|
**|
|
|
|
|
|
|
|
|
|
|
|
|
|
0.000
0.057
0.303
0.060
0.002
0.084
0.063
0.395
0.038
0.023
0.172
0.013
0.108
420
The REG Procedure
Model: MODEL1
R-Square Selection Method
Regression Models for Dependent Variable: y
Number in
Model
R-Square
AIC
This output was produced by the
option
It indicates that all five regression
Variables
in Model
SBC
e
in the model statement of the GLM procedure.
parameters are estimable.
1
0.6744
58.8383
59.96815 x4
1
0.6661
59.1672
60.29712 x2
1
0.5342
63.4964
64.62630 x1
1
0.2860
69.0481
70.17804 x3
-----------------------------------------------------2
0.9787
25.3830
27.07785 x1 x2
2
0.9726
28.6828
30.37766 x1 x4
2
0.9353
39.8308
41.52565 x3 x4
2
0.8470
51.0247
52.71951 x2 x3
2
0.6799
60.6172
62.31201 x2 x4
2
0.5484
65.0933
66.78816 x1 x3
------------------------------------------------------3
0.9824
24.9187
27.17852 x1 x2 x4
3
0.9823
24.9676
27.22742 x1 x2 x3
3
0.9814
25.6553
27.91511 x1 x3 x4
3
0.9730
30.4953
32.75514 x2 x3 x4
------------------------------------------------------4
0.9824
26.8933
29.71808 x1 x2 x3 x4
The GLM Procedure
General Form of Estimable Functions
Effect
Coefficients
Intercept
L1
x1
L2
x2
L3
x3
L4
x4
L5
422
421
This output was produced by the
e1
option
in the model statement of the GLM procedure.
It describes the null hypotheses that are
Type II Estimable Functions
tested with the sequential Type I sums of
squares.
----Coefficients---Effect
Type I Estimable Functions
x1
x2
x3
x4
Intercept
0
0
0
0
----------------Coefficients---------------x1
x2
x3
x4
x1
L2
0
0
0
Effect
Intercept
0
0
0
0
x2
0
L3
0
0
x1
L2
0
0
0
x3
0
0
L4
0
x2
0.6047*L2
L3
0
0
x3
-0.8974*L2
0.0213*L3
L4
0
x4
0
0
0
L5
x4
-0.6984*L2
-1.0406*L3
-1.0281*L4
L5
423
424
>
>
>
>
>
>
# The commands are stored in:
#
#
#
#
cement.spl
The data file is stored under the name
cement.dat. It has variable names on the
first line. We will enter the data into
a data frame.
> cement <- read.table("cement.txt", header=T)
> cement
1
2
3
4
5
6
7
8
9
10
11
12
13
run
1
2
3
4
5
6
7
8
9
10
11
12
13
X1
7
1
11
11
7
11
3
1
2
21
1
11
10
> #
Compute correlations and round the results
> #
to four significant digits
> round(cor(cement[-1]),4)
X2
26
29
56
31
52
55
71
31
54
47
40
66
68
X3
6
15
8
8
6
9
17
22
18
4
23
9
8
X4
60
52
20
47
33
22
6
44
22
26
34
12
12
Y
78.5
74.3
104.3
87.6
95.9
109.2
102.7
72.5
93.1
115.9
83.8
113.2
109.4
X4
Y
X1
1.0000
X1
0.2286 -0.8241 -0.2454
X2
X3
0.7309
X2
0.2286
1.0000 -0.1392 -0.9730
0.8162
X3 -0.8241 -0.1392
1.0000
0.0295 -0.5348
X4 -0.2454 -0.9730
0.0295
1.0000 -0.8212
Y
0.7309
0.8162 -0.5348 -0.8212
1.0000
426
425
10
30
50
15
20
30 40 50 60 70
> #
to open a graphics wundow
should first use motif( )
X2
40
Unix users
60
Create a scatterplot matrix with smooth
curves.
50
> #
> #
70
5
10
X1
30
>
15
10
5
}
X4
pairs(cement[ ,-1], panel=points.lines)
110
>
100
par(din=c(7,7), pch=18, mkh=.15, cex=1.2, lwd=3)
10
>
20
30
>
Y
80
90
lines(loess.smooth(x, y, 0.90))
+
60
+
X3
50
points(x, y)
40
{
+
20
> points.lines <- function(x, y)
+
5
427
10
15
20
5
10
15
20
80 90
110
428
> #
Fit a
> #
and Ripley, Chapter 6)
linear regression model (Venables
Correlation of Coefficients:
(Intercept)
> cement.out <- lm(Y~X1+X2+X3+X4, cement)
Call: lm(formula = Y ~ X1+X2+X3+X4, data=cement)
Residuals:
Min
1Q Median
3Q
X2
X3
X1 -0.9678
> summary(cement.out)
-3.176 -1.674
X1
X2 -0.9978
0.9510
X3 -0.9769
0.9861
0.9624
X4 -0.9983
0.9568
0.9979
0.9659
Max
0.264 1.378 3.936
> anova(cement.out)
Coefficients:
Value
Std. Error
t value
Pr(>|t|)
(Intercept)
63.1660
69.9338
0.9032
0.3928
X1
1.5431
0.7433
2.0759
0.0716
X2
0.5020
0.7224
0.6949
0.5068
X3
0.0942
0.7532
0.1250
0.9036
X4
-0.1515
0.7077
-0.2141
0.8358
Analysis of Variance Table
Response: Y
Terms added sequentially (first to last)
F Value
Pr(F)
X1
1
1448.754 1448.754 243.0978
0.0000
Residual standard error: 2.441 on 8 degrees of freedom
X2
1
1205.703 1205.703 202.3144
0.0000
Multiple R-Squared: 0.9824
X3
1
9.790
9.790
1.6428
0.2358
X4
1
0.273
0.273
0.0458
0.8358
Residuals
8
47.676
5.960
F-statistic: 111.8 on 4 and 8 degrees of freedom,
Df Sum of Sq
Mean Sq
the p-value is 4.707e-007
430
429
> #
Create a function to evaluate an orthogonal
> #
projection matrix.
Then create a function
> #
to compute type II sums of squares.
> #
This uses the ginv( )
> #
library, so you must attach the MASS library
function in the MASS
> library(MASS)
> #=======================================
> # project( )
> #-------------> # calculate orthogonal projection matrix
> #=======================================
> project <- function(X)
+
{ X%*%ginv(crossprod(X))%*%t(X)
}
> #=======================================
431
>
>
>
>
>
>
>
>
>
>
>
+
+
+
+
+
+
+
+
+
+
+
+
+
+
#========================================
# typeII.SS( )
#-----------------# calculate Type II sum of squares
#
# input lmout = object made by the
#
lm( ) function
#
y = dependent variable
#========================================
typeII.SS <- function(lmout,y)
{
# generate the model matrix
model <- model.matrix(lmout)
# create list of parameter names
par.name <- dimnames(model)[[2]]
# compute number of parameters
n.par <- dim(model)[2]
# Compute residual mean square
SS.res <- deviance(lmout)
df2
<- lmout$df.resid
MS.res <- SS.res/df2
432
+
result <- NULL
# store results
+
+
# Compute Type II SS
+
for (i in 1:n.par) {
+
A <- project(model)-project(model[,-i])
+
SS.II
<- t(y) %*% A %*% y
+
df1
<- qr(project(model))$rank -
+
> typeII.SS(cement.out, cement$Y)
qr(project(model[ ,-i]))$rank
+
MS.II
+
F.stat <- MS.II/MS.res
+
p.val
<- 1-pf(F.stat,df1,df2)
+
temp
<- cbind(df1,SS.II,MS.II,F.stat,p.val)
+
<- SS.II/df1
Analysis of Variance (TypeII Sum of Squares)
Df Sum of Sq
(Inter.)
result <- rbind(result,temp)
+
}
+
+ result<-rbind(result,c(df2,SS.res,MS.res,NA,NA))
+ dimnames(result)<-list(c(par.name,"Residual"),
+ c("Df","Sum of Sq","Mean Sq","F Value","Pr(F)"))
1
4.861907
Mean Sq
F Value
Pr(F)
4.861907 0.815818 0.392790
X1
1 25.682254 25.682254 4.309427 0.071568
X2
1
2.878010
2.878010 0.482924 0.506779
X3
1
0.093191
0.093191 0.015637 0.903570
1
0.273229
0.273229 0.045847 0.835810
X4
Residual
8 47.676412
5.959551
NA
NA
+ cat("Analysis of Variance (TypeII Sum of Squares)
+
+
\n")
round(result,6)
+ }
> #==========================================
433
434
> # Venables and Ripley have supplied functions
> # studres( ) and stdres( ) to compute
Response Predicted Residual Stud. Res. Std. Res.
> # studentized and standardized residuals.
1
78.5
78.4929
0.0071
0.0040
> # You must attach the MASS library before
2
74.3
72.8005
1.4995
0.7299
0.0043
0.7522
> # using these functions.
3
104.3
105.9744
-1.6744
-1.0630
-1.0545
4
87.6
89.3333
-1.7333
-0.8291
-0.8458
0.1349
> cement.res <- cbind(cement$Y,cement.out$fitted,
5
95.9
95.6360
0.2640
0.1264
+
cement.out$resid,
6
109.2
105.2635
3.9365
2.0324
1.7230
+
studres(cement.out),
7
102.7
104.1289
-1.4289
-0.7128
-0.7358
+
stdres(cement.out))
8
72.5
75.6760
-3.1760
-1.9745
-1.6917
9
93.1
91.7218
1.3782
0.6472
0.6721
> dimnames(cement.res) <- list(cement$run,
10
115.9
115.6010
0.2990
0.2100
0.2237
+
11
83.8
81.8034
1.9966
1.0919
1.0790
12
113.2
112.3007
0.8993
0.4061
0.4291
13
109.4
111.6675
-2.2675
-1.1326
-1.1131
+
c("Response","Predicted","Residual",
"Stud. Res.","Std. Res."))
> round(cement.res,4)
435
436
> # Produce plots for model diagnostics including
> # Cook's D.
Unix users should first use motif()
> # to open a
graphics window
> par(mfrow=c(3,2))
> plot(cement.out)
> # Search for a simpler model
> cement.stp <- step(cement.out,
+
scope=list(upper = ~X1 + X2 + X3 + X4,
+
lower = ~ 1), trace=F)
438
2.0
4
437
6
6
1.5
1.0
13
0.5
0
-2
Residuals
2
sqrt(abs(Residuals))
8
> cement.stp$anova
13
8
80
90
100
110
80
90
Fitted : X1 + X2 + X3 + X4
100
110
Stepwise Model Path
fits
4
Analysis of Deviance Table
110
6
0
Y ~ X1 + X2 + X3 + X4
-2
80
90
Y
Residuals
100
2
Initial Model:
Final Model:
13
8
80
90
100
110
-1
Fitted : X1 + X2 + X3 + X4
0.4
0.3
0.2
f-value
Step Df Deviance Resid. Df Resid. Dev
1
0.1
11
AIC
8
47.67641 107.2719
2 - X3
1 0.093191
9
47.76960
95.4460
3 - X4
1 9.970363
10
57.73997
93.4973
0.0
-20
0.6
8
3
0.2
20
10
0
-10
Cook’s Distance
20
Residuals
10
-10
-20
0.2
Y ~ X1 + X2
1
Quantiles of Standard Normal
Y
0
Fitted Values
0
0.6
2
4
6
8
10
12
Index
439