Lab 5 Key
advertisement

Lab 5 Key
#Here is some code for Lab #5
#First we study prediction and tolerance intervals
# 1 simulate 10000 sets of observations
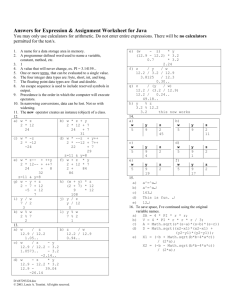
M<-matrix(rnorm(60000,mean=5,sd=1.715),nrow=10000,byrow=T)
av<-1:10000
s<-1:10000
Lowpi<-rep(0,10000)
Uppi<-rep(0,10000)
Lowti<-rep(0,10000)
Upti<-rep(0,10000)
chkpi<-rep(0,10000)
chkti<-rep(0,10000)
for (i in 1:10000){
av[i]<-mean(M[i,1:5])
}
for (i in 1:10000){
s[i]<-sd(M[i,1:5])
}
# if the interval contain 6th col, chkpi is an indicator for this
for(i in 1:10000) {Lowpi[i]<-av[i]-2.132*s[i]*sqrt(1+(1/5))}
for(i in 1:10000) {Uppi[i]<-av[i]+2.132*s[i]*sqrt(1+(1/5))}
for(i in 1:10000) {if((Lowpi[i]<M[i,6])&(M[i,6]<Uppi[i])) chkpi[i]<-1}
#check the first 10 rows to have a look
cbind(Lowpi[1:10],Uppi[1:10],M[1:10,6],chkpi[1:10])
##
## [1,]
## [2,]
## [3,]
## [4,]
## [5,]
## [6,]
## [7,]
## [8,]
## [9,]
## [10,]
[,1]
0.0013140
-1.4275700
1.5367144
2.3205625
0.1701937
5.0471106
-1.2846655
-2.6656065
1.4062821
-0.6686512
[,2]
10.191154
10.033024
10.844678
10.124524
9.286392
8.337752
12.181363
12.882861
6.278091
8.885316
[,3] [,4]
2.406896
1
9.642363
1
7.551604
1
5.021665
1
3.409563
1
4.187643
0
5.551324
1
4.395546
1
2.881683
1
3.158086
1
mean(chkpi)
## [1] 0.8975
#6.655 in page 800 99% percent significant confidence
for(i in 1:10000) {Lowti[i]<-av[i]-6.655*s[i]}
for(i in 1:10000) {Upti[i]<-av[i]+6.655*s[i]}
for(i in 1:10000) {if(pnorm(Upti[i],mean=5,sd=1.715)-pnorm(Lowti[i],mean=5,sd
=1.715)>.9) chkti[i]<-1}
cbind(Lowti[1:10],Uppi[1:10],pnorm(Upti[1:10],mean=5,sd=1.715)-pnorm(Lowti[1:
10],mean=5,sd=1.715),chkti[1:10])
##
[,1]
[,2]
[,3] [,4]
## [1,] -9.421793 10.191154 1.000000
1
## [2,] -12.025814 10.033024 1.000000
1
## [3,] -7.070873 10.844678 1.000000
1
## [4,] -4.896191 10.124524 1.000000
1
## [5,] -8.260057 9.286392 1.000000
1
## [6,]
2.004073 8.337752 0.959573
1
## [7,] -13.737445 12.181363 1.000000
1
## [8,] -17.044132 12.882861 1.000000
1
## [9,] -3.098948 6.278091 0.999626
1
## [10,] -9.503732 8.885316 1.000000
1
mean(chkti)
## [1] 0.9901
#2.Now we plot some Chi-square densities
curve(dchisq(x,df=2),xlim=c(0,25))
curve(dchisq(x,df=4),add=TRUE,xlim=c(0,25))
curve(dchisq(x,df=6),add=TRUE,xlim=c(0,25))
curve(dchisq(x,df=10),add=TRUE,xlim=c(0,25))
#Now we plot some F densities: ratio of sample variance
curve(df(x,df1=20,df2=20),xlim=c(0,10))
curve(df(x,df1=3,df2=20),add=TRUE,xlim=c(0,10))
curve(df(x,df1=20,df2=3),add=TRUE,xlim=c(0,10))
curve(df(x,df1=3,df2=3),add=TRUE,xlim=c(0,10)) #less compact for smaller degr
ees of freedom
#3 Here's a bit of a "robustness" study for a t CI for a mean
M<-matrix(runif(50000,min=0,max=1),nrow=10000,byrow=T)
av<-1:10000
s<-1:10000
Lowci<-rep(0,10000)
Upci<-rep(0,10000)
chkci<-rep(0,10000)
for (i in 1:10000){
av[i]<-mean(M[i,1:5])
}
for (i in 1:10000){
s[i]<-sd(M[i,1:5])
}
for(i in 1:10000) {Lowci[i]<-av[i]-2.132*s[i]/sqrt(5)}
for(i in 1:10000) {Upci[i]<-av[i]+2.132*s[i]/sqrt(5)}
for(i in 1:10000) {if((Lowci[i]<.5)&(.5<Upci[i])) chkci[i]<-1}
cbind(Lowci[1:10],Upci[1:10],chkci[1:10])
##
## [1,]
## [2,]
## [3,]
## [4,]
## [5,]
## [6,]
## [7,]
## [8,]
## [9,]
## [10,]
[,1]
0.1025537
0.2483314
0.1713004
0.2757000
0.2587950
0.4132108
0.2513104
0.1872738
0.3043536
0.4468239
[,2] [,3]
0.8621661
1
0.8382700
1
0.6182579
1
0.7166218
1
0.7636928
1
0.9118333
1
0.6510614
1
0.7808194
1
0.7132618
1
0.9425616
1
mean(chkci)
## [1] 0.8863
##Here is a bit of a study on Exponential distribution
M<-matrix(rexp(50000,rate=1),nrow=10000,byrow=T)
av<-1:10000
s<-1:10000
Lowci<-rep(0,10000)
Upci<-rep(0,10000)
chkci<-rep(0,10000)
for (i in 1:10000){
av[i]<-mean(M[i,1:5])
}
for (i in 1:10000){
s[i]<-sd(M[i,1:5])
}
for(i in 1:10000) {Lowci[i]<-av[i]-2.132*s[i]/sqrt(5)}
for(i in 1:10000) {Upci[i]<-av[i]+2.132*s[i]/sqrt(5)}
for(i in 1:10000) {if((Lowci[i]<.5)&(.5<Upci[i])) chkci[i]<-1}
cbind(Lowci[1:10],Upci[1:10],chkci[1:10])
##
##
##
##
##
##
##
##
##
[,1]
[,2] [,3]
[1,] 0.35068938 1.6319212
1
[2,] -0.07203980 0.8563114
1
[3,] -0.24119922 1.7939243
1
[4,] 0.47712501 1.6061637
1
[5,] 0.35008052 1.6271238
1
[6,] 0.54408786 2.4953199
0
[7,] 0.10979705 2.1101664
1
[8,] 0.26327599 1.2129589
1
## [9,]
## [10,]
0.05996656 2.9856460
0.04114604 1.1973431
1
1
mean(chkci)
## [1] 0.8593
#Here is a bit of a study on the effectiveness of CIs for p
p<-.2
n<-1600
X<-rbinom(10000,size=n,p)
phat<-X/n
phattilde<-(X+2)/(n+4)
Lowcip<-1:10000
Upcip<-1:10000
Lowciptilde<-1:10000
Upciptilde<-1:10000
chkcip<-rep(0,10000)
chkciptilde<-rep(0,10000)
#chk for phat
for(i in 1:10000) {Lowcip[i]<-phat[i]-1.645*sqrt(phat[i]*(1-phat[i])/n)}
for(i in 1:10000) {Upcip[i]<-phat[i]+1.645*sqrt(phat[i]*(1-phat[i])/n)}
for(i in 1:10000) {if((Lowcip[i]<p)&(p<Upcip[i])) chkcip[i]<-1}
#chk for phattilde
for(i in 1:10000) {Lowciptilde[i]<-phat[i]-1.645*sqrt(phattilde[i]*(1-phattil
de[i])/n)}
for(i in 1:10000) {Upciptilde[i]<-phat[i]+1.645*sqrt(phattilde[i]*(1-phattild
e[i])/n)}
for(i in 1:10000) {if((Lowciptilde[i]<p)&(p<Upciptilde[i])) chkciptilde[i]<-1
}
mean(chkcip)
## [1] 0.9004
mean(chkciptilde)
## [1] 0.9004
#for n=50,p=0.01 we have mean(chkcip)[1] 0.3978 mean(chkciptilde)0.9998
#for n=50,p=0.001 we have mean(chkcip)[1] 0.0506 mean(chkciptilde)1
#for n=1600,p=0.001 we have mean(chkcip)[1] 0.7859 mean(chkciptilde)0.9988
#for n=1600,p=0.001 we have mean(chkcip)[1] 0.8974 mean(chkciptilde)0.8974
#From above we can see that increasing the sample size these two methods work
the same, while with small p and sample size, the tilde method is better than
that of cip.
#Here is a quick way to make a single normal plot
#(There is code for making multiple normal plots on a single set of axes in t
he HW)
qqnorm(c(1,4,3,6,8,34,3,6,2,7),datax=TRUE)
#Here is some R code for the class "Depth of Cut" example
pulses<-c(rep("A(100)",4),rep("B(500)",4),rep("C(1000)",4))
depth<-c(7.4,8.6,5.6,8.0,24.2,29.5,26.5,23.8,33.4,37.5,35.9,34.8)
Depth<-data.frame(depth,pulses)
Depth
##
## 1
## 2
depth
7.4
8.6
pulses
A(100)
A(100)
##
##
##
##
##
##
##
##
##
##
3
4
5
6
7
8
9
10
11
12
5.6
8.0
24.2
29.5
26.5
23.8
33.4
37.5
35.9
34.8
A(100)
A(100)
B(500)
B(500)
B(500)
B(500)
C(1000)
C(1000)
C(1000)
C(1000)
plot(depth ~ pulses,data=Depth)
plot(as.factor(pulses),depth) #Only loss of the labels
summary(Depth)
##
##
##
##
##
##
##
depth
Min.
: 5.60
1st Qu.: 8.45
Median :25.35
Mean
:22.93
3rd Qu.:33.75
Max.
:37.50
pulses
A(100) :4
B(500) :4
C(1000):4
aggregate(Depth$depth,by=list(Depth$pulses),mean)
##
Group.1
x
## 1 A(100) 7.4
## 2 B(500) 26.0
## 3 C(1000) 35.4
aggregate(Depth$depth,by=list(Depth$pulses),sd)
##
Group.1
x
## 1 A(100) 1.296148
## 2 B(500) 2.619160
## 3 C(1000) 1.733974
depth.aov<-aov(depth ~ pulses,data=Depth)
summary(depth.aov)
##
##
##
##
##
Df Sum Sq Mean Sq F value
Pr(>F)
pulses
2 1624.4
812.2
211 2.75e-08 ***
Residuals
9
34.6
3.8
--Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1