Reshma P. Shetty†‡ and Thomas F. Knight, Jr.‡
advertisement

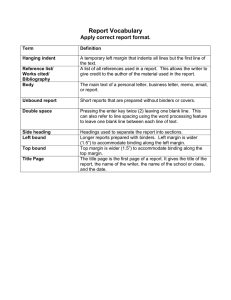
Reshma P. Shetty†‡ and Thomas F. Knight, Jr.‡ †Biological Engineering Division, ‡Computer Science and Artificial Intelligence Laboratory, MIT Model formulation α sensitivity analysis i 8 K1=αi*100 and K2=αi/10000 7 Noise margin (%) 6 3 0 2 10 3 4 10 5 10 10 αi (proteins/cell) K1 sensitivity analysis K2 sensitivity analysis 8 8 α =49561 and K =α /10000 i Figure 3: The swing, noise margin and trip point are quantitative measures of the quality of the transfer characteristic. Ideal devices maximize the noise margin and have a trip point close to half the device swing. Transfer curve of C0051, R0051 (λ) Geometric mean fluorescence (AU) Geometric mean fluorescence (AU) 20 15 10 5 0 0 10 1 2 10 10 3 10 250 200 150 100 50 0 0 10 1 [IPTG] uM Geometric mean fluorescence (AU) 1400 1200 1000 800 600 400 200 1 3 10 2 10 i 7 6 6 5 4 3 4 3 2 1 1 1 2 10 3 10 4 5 10 K (proteins/cell) 10 6 10 0 0 10 7 10 1 10 1 K1K2 So = K1K2 + [pi]2 Figure 6: Key parameters that affect device behavior are the steady-state maximum protein level αi, promoter copy number [do]T and dissociation constants K1 and K2. i 5 2 10 1 2 10 3 10 K (proteins/cell) 4 10 5 10 6 10 2 Figure 8: Noise margin as a function of parameter value for αi, K1 and K2. Device performance is sensitive to K1 and K2 but not αi. Alternative design Transfer curve of C0053, R0053 (P22) 1600 10 10 [IPTG] uM Transfer curve of C0052, R0052 (434) 0 0 10 2 10 α =49561 and K =α *100 i 7 0 0 10 T 2 ] [p ] 2[d o i T −1 2 [pi] =Siαi = [pi] + 2K1 [pi] + K1K2 + [pi]2 kspiksmi T αi ≡ [di] kdpikdmi Existing devices fail 25 Model equations 2 Noise margin (%) Figure 5: Model includes synthesis, degradation and binding reactions. 3 10 150 Target parameter values 100 Inverter transfer curve 1 S vs S 50 o i S vs S 0.9 i o 0.8 0 0 10 [IPTG] uM Figure 1: An inverter is a simple digital logic device. The work presented here focuses on characterizing and modeling a transcriptional inverter. 4 1 Geometric mean fluorescence (AU) Implement in vivo combinational digital logic using transcription-based devices. 5 2 Transfer curve of C0050, R0050 (HK022) Goal Sensitivity analysis Noise margin (%) The goal of Synthetic Biology is to engineer systems from biological parts. One class of systems are those whose purpose is to process information. My work seeks to build transcription-based devices for use in combinational digital logic. Preliminary characterization experiments show that existing devices fall short of desired device behavior. I propose to develop a novel implementation of transcription-based logic by designing synthetic transcription factors from well-characterized DNA binding and dimerization domains. Initial modeling work serves to inform design of these devices. Device performance 1 2 10 10 So (blue) or Si (green) Abstract 3 10 [IPTG] uM Figure 4: Experimentally-determined transfer characteristics for 4 transcriptional inverter prototypes. 0.7 N =0.05 0.6 SM =0.2 0.5 0.4 0.3 Figure 9: A possible alternate device design includes additional nonfunctional protein binding sites on the DNA. 0.2 0.1 0 0 0.2 0.4 0.6 Si (blue) or So (green) 0.8 1 Inverter transfer curve So vs Si Figure 7: These parameter values lead to acceptable device behavior. K1 ≈ αi ∗ 100 ≈ 10 mM (dimerization) K2 ≈ αi/10000 ≈ 10 nM (DNA binding) (assuming αi ≈ 50, 000 proteins/cell) 0.8 0.7 N =0.1 =0.05 0.6 S =0.4 =0.2 Modeling informs design of synthetic transcription factors for digital logic. Simplifying assumptions Figure 2: A transfer characteristic is a plot of device output as a function of device input. It describes static device performance. 1. Use reaction rate equations 2. Neglect cell growth and DNA replication 3. Ignore details of synthesis and degradation reactions 4. All or none repression 5. Binding reactions at equilibrium 6. Steady-state • Steady-state maximum input protein concentration αi is the input protein swing. • Value of dissociation constants relative to αi determines noise margin. • Inverter skew may be a problem. References [1] G. K. Ackers, A. D. Johnson, and M. A. Shea. Quantitative model for gene regulation by λ phage repressor. Proc Natl Acad Sci USA, 79(4):1129–33, 1982. [2] M. A. Gibson and E. Mjolsness. Computational Modeling of Genetic and Biochemical Networks, chapter 1, pages 3–48. The MIT Press, Cambridge, Massachusetts, 2001. [3] C. F. Hill. Noise margin and noise immunity in logic circuits. Microelectronics, 1:16–22, April 1968. [4] R. Weiss, T. F. Knight Jr, and G. Sussman. Cellular Computing, chapter 4, pages 43–73. Series in Systems Biology. Oxford University Press, 2004. M 0.5 0.4 0.3 0.2 0.1 0 0 Device behavior Si vs So 0.9 So (blue) or Si (green) To engineer good devices, we need device performance metrics and measurement methods. One inverter has good performance characteristics; however, the other devices fail to function properly. 1 0.2 0.4 0.6 Si (blue) or So (green) 0.8 1 [da]T = 0.1αi and K6 = K2/10 Figure 10: This alternate design can lead to enhanced device performance. Alternate device designs may lead to improved transfer characteristics. Acknowledgements I would like to thank Tom Knight, Drew Endy, Austin Che, Scot Wolfe, Keith Joung, Bruce Tidor, Bob Sauer, Bambang Adiwijaya, the Knight lab and the MIT Synthetic Biology Working Group for valuable discussions and support. Funding for this work has been generously provided by the NSF Graduate Research Fellowship, the Whitaker Foundation Graduate Research Fellowship and the Andy and Erna Viterbi Graduate Fellowship in Computational Biology.