Supplementary Table 2. Cis-acting elements analysis of 5'
advertisement

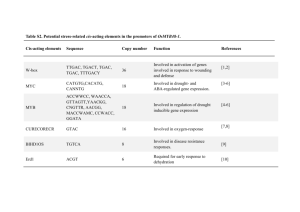
Supplementary Table 2. Cis-acting elements analysis of 5'-upstream regulatory elements and their positions in bi-directional promoter (672 bp) identified by PLACE and PLANTCARE. Name of element ABRE the Signal Sequence ACE AAACGTTA Box-4 ATTAAT Box-1 Box-III Box-W1 TTTCAAA CATTTACACT TTGACC Position relative Functional description to ATG +137 Cis-acting element involved in the abscisic acid responsiveness. +164 Cis-acting element involved in light responsiveness. +329 Part of a conserved DNA module involved in light responsiveness. -345 Light responsive element. -552 Protein binding site. +605, -617 Fungal elicitor responsive element. CATT motif CGTCA-motif GCATTC CGTCA +216 +569, -224 ERE ATTTCAAA -345 G-Box CACGTA -137 G-Box GTACGTG TACGTG CACGTC +136, +137, -225 GA-motif AAAGATGA -12 Part of a light responsive element. Skn-1-motif GTCAT +477 Sp1 TC-rich repeats CC(G/A)CCC GTTTTCTTAC -357 +661 TGACG-motif TGACG +224, -569 W box TTGACC +605, -617 Cis-acting regulatory element required for endosperm expression. Light responsive element. Cis-acting element involved in defense and stress responsiveness. Cis-acting regulatory element involved in the MeJAresponsiveness. - WUN-motif TCATTACGAA -627 Wound-responsive element. as-2-box GATAatGATG -1, -7, -4 Involved in shoot-specific expression and light responsiveness. 2SSEEDPROTB ANAPA CAAACAC +500, -130 Conserved in many storage-protein gene promoters; May be important for high activity of the napA promoter. TACGTG Part of a light responsive element. Cis-acting regulatory element involved in the MeJAresponsiveness. Ethylene-responsive element. Cis-acting regulatory element involved in light responsiveness. Cis-acting regulatory element involved in light responsiveness. Organism Arabidopsis thaliana Petroselinum crispum Petroselinum crispum Pisum sativum Pisum sativum Petroselinum crispum Zea mays Hordeum vulgare Dianthus caryophyllus Antirrhinum majus Oryza sativa, Daucus carota, Zea mays Helianthus annuus Oryza sativa Zea mays Nicotiana tabacum Hordeum vulgare Arabidopsis thaliana Brassica oleracea Nicotiana tabacum Brassica napus ABRELATERD1 ACGTG +138 ACGTABREMO TIFA2OSEM ACGTGKC +226 ACGTATERD1 ACGT +138, +167, +226 +574, -138, -167, -226, -574 ACGTOSGLUB1 GTACGTG +136 ACGTTBOX AACGTT + 166, -166 AMYBOX1 TAACARA -305 ARR1AT NGATT +315, -25, -275 ASF1MOTIFCA MV TGACG +224, -569 CACTFTPPCA1 YACT +68, 66, +90, 453, +108, -539, +310, -542, +483, -552, +654 -648 CANBNNAPA CNAACAC +500, +130 ABRE-like sequence (from -199 to 195) required for etiolation-induced expression of erd1 (early responsive to dehydration) in Arabidopsis Experimentally determined sequence requirement of ACGT-core of motif A in ABRE of the rice gene, DRE and ABRE are interdependent in the ABA-responsive expression of the rd29A. Arabidopsis thaliana ACGT sequence (from -155 to -152) required for etiolation-induced expression of erd1 (early responsive to dehydration) in Arabidopsis ACGT motif" found in GluB-1 gene in rice, Required for endospermspecific expression; Conserved in the 5'-flanking region of glutelin genes. Arabidopsis thaliana Plant bZIP Proteins gather at ACGT elements. Conserved sequence found in 5'upstream region of alpha-amylase gene. Plant "ARR1-binding element" found in Arabidopsis; ARR1 is a response regulator; N=G/A/C/T, AGATT is found in the promoter of rice nonsymbiotic haemoglobin-2 gene. TGACG motifs are found in many promoters and are involved in transcriptional activation of several genes by auxin and/or salicylic acid. (CACT) is a key component of Mem1 (mesophyll expression module 1) found in the Cisregulatory element in the distal region of the phosphoenolpyruvate carboxylase (ppcA1) of the C4 dicot F. trinervia. Core of "(CA)n element" in storage protein genes in Brassica napus, Embryo and endosperm-specific transcription of napA gene, seed Oryza sativa, Arabidopsis thaliana Oryza sativa Hordeum vulgare, Oryza sativa, Triticum aestivum Arabidopsis thaliana Cauliflower mosaic virus, Nicotiana Tabacum, Arabidopsis thaliana Flaveria trinervia Brassica napus specificity; activator and repressor. A variant of CArG motif, with a longer A/T-rich core; Binding site for AGL15. Common sequence found in the 5'non-coding regions of eukaryotic genes. The sequence critical for cytokininenhanced Protein Binding in vitro, found in -490 to -340 of the promoter of the cucumber POR (NADPHprotochlorophyllide reductase) gene. GTAC is the core of a CuRE (copper-response element) found in Cyc6 and Cpx1 genes in Chlamydomonas, also involved in oxygen-response of these genes. Core site required for binding of Dof proteins in maize. CARGCW8GAT CWWWWWWW WG + 205, + 328, 205, -328 Arabidopsis thaliana CCAATBOX1 CCAAT + 341, -73 CPBCSPOR TATTAG +62 CURECORECR GTAC +67, +89, +136, 67, -89, -136 DOFCOREZM AAAG +300, -16, +379, 174, +523, -221, +533, -405, 485, -603 DPBFCOREDC DC3 ACACNNG -550 A novel class of bZIP transcription factors, DPBF-1 and 2 (Dc3 promoter-binding factor-1 and 2) binding core sequence. Daucus carota, Arabidopsis thaliana EBOXBNNAPA CANNTG +550, -550 Brassica napus EECCRCAH1 GANTTNC +104 ELRECOREPCR P1 TTGACC +605, -617 ERELEE4 AWTTCAAA -345 E-box of napA storage-protein gene of Brassica napus, (also known as Rresponse element). "EEC"; Consensus motif of the two enhancer elements, EE-1 and EE-2, both found in the promoter region of the Chlamydomonas Cah1 (encoding a periplasmic carbonic anhydrase); Binding site of Myb transcription factor LCR1. ElRE (Elicitor Responsive Element) core of parsley PR1 genes; consensus sequence of elements W1 and W2 of parsley PR1-1 and PR1-2 promoters; Box W1 and W2 are the binding site of WRKY1 and WRKY2, respectively. "ERE (ethylene responsive element)" of tomato E4 and carnation GST1 genes. ERE motifs mediate ethyleneinduced activation of the U3 promoter region. Eukaryotes,,Gl ycine max Cucumis sativus Chlamydomon as reinhardtii Zea mays Chlamydomon as reinhardtii Petroselinum crispum); Nicotiana tabacum Lycopersicon esculentum, Dianthus caryophyllus, Lycopersicon chilense GADOWNAT ACGTGTC +226 GARE1OSREP1 TAACAGA -305 GT1CONSENSU S GRWAAW +76, +390, +469, -17, -242, -264 GTGANTG10 GTGA +142 HEXMOTIFTAH ACGTCA 3H4 (-) 224 IBOX GATAAG +449 IBOXCORE GATAA +390, +449, +469, -243 Sequence present in 24 genes in the GA-down regulated d1 cluster (106 genes) found in Arabidopsis seed germination; This motif is similar to ABRE. "Gibberellin-responsive element (GARE)" found in the promoter region of a cystein proteinase (REP1) gene in rice. Consensus GT-1 binding site in many light-regulated genes, e.g., RBCS from many species, PHYA from oat and rice, spinach RCA and PETA, and bean CHS15; R=A/G; W=A/T .GT-1 can stabilize the TFIIA-TBP-DNA (TATA box) complex; Binding of GT-1-like factors to the PR-1a promoter influences the level of SA-inducible gene expression. "GTGA motif" found in the promoter of the tobacco late pollen gene g10 which shows homology to pectate lyase and is the putative homologue of the tomato gene lat56; Located between -96 and -93. Arabidopsis thaliana "Hexamer motif" found in promoter of wheat histone genes H3 and H4; CaMV35S; NOS; Binding with HBP1A and HBP-1B; Binding site of wheat nuclear protein HBP-1 (histone DNA binding protein-1); HBP-1 has a leucine zipper motif; "hexamer motif" in type 1 element may play important roles in regulation of replication- dependent but not of replication-independent expression of the wheat histone H3 gene; Rice OBF1-homodimerbinding site. Conserved sequence upstream of light-regulated genes; Sequence found in the promoter region of rbcS of tomato and Arabidopsis; Binding site of LeMYB1, (transcriptional activator) that is a member of a novel class of myb-like proteins. Conserved sequence upstream of light-regulated genes. Triticum aestivum, CaMV, Oryza sativa Oryza sativa Pisum sativum, Avena sativa, Oryza sativa, Nicotiana tabacum, Arabidopsis thaliana, Spinacia oleracea, bean Triticum aestivum, CaMV, Oryza sativa Lycopersicon esculentum, Arabidopsis thaliana Monocots and dicots. INRNTPSADB YTCANTYY +177 LECPLEACS2 TAAAATAT +392 MYB1AT WAACCA +324 MYBCORE CNGTTR +306 MYBST1 GGATA -643, -650 MYCCONSENS USAT CANNTG +550, -550 NAPINMOTIFB N TACACAT -420 NODCON1GM and OSE1ROOTNO DULE AAAGAT +300, -14, -601 NODCON2GM and OSE2ROOTNO CTCTT +564 Initiator elements found in the tobacco psaDb gene promoter without TATA boxes; Lightresponsive transcription of psaDb depends on Inr, but not TATA box. Core element in LeCp (tomato Cys protease) binding cis-element (from 715 to -675) in LeAcs2 gene. Nicotiana tabacum MYB recognition site found in the promoters of the dehydrationresponsive gene rd22 and many other genes in Arabidopsis. Binding site for all animal MYB and at least two plant MYB proteins ATMYB1 and ATMYB2, both; ATMYB2 is involved in regulation of genes that are responsive to water stress in Arabidopsis; A petunia MYB protein (MYB.Ph3) is involved in regulation of flavonoid biosynthesis. Core motif of MybSt1 (a potato MYB homolog) binding site. MYC recognition site found in the promoters of the dehydrationresponsive gene rd22 and many other genes in Arabidopsis; Binding site of ATMYC2 (previously known as rd22BP1); (E-box; CANNTG), (MYCATRD22); N=A/T/G/C; MYC recognition sequence in CBF3 promoter; Binding site of ICE1 (inducer of CBF expression 1) that regulates the transcription of CBF/DREB1 genes in the cold in Arabidopsis. Sequence found in 5' upstream region (-6, -95, -188) of napin gene in Brassica napus; Interact with a protein present in crude nuclear extracts from developing seeds. One of two putative nodulin consensus sequences. Arabidopsis thaliana One of two putative nodulin consensus sequences. Glycine max Lycopersicon esculentum Arabidopsis thaliana, animal, Petunia hybrida Solanum tuberosum Arabidopsis thaliana Brassica napus Glycine max DULE NRRBNEXTA TAGTGGAT - 651 POLASIG1 AATAAA + 296, + 429, +529, -60 POLASIG2 AATTAAA +149 POLASIG3 AATAAT +579 POLLEN1LELA T52 AGAAA PRECONSCRHS P70A "NRR (negative regulatory region)" in promoter region of Brassica napus extA gene; Removal of this region leads to expression in all tissues within the stem internode, petiole and root. PolyA signal found in legA gene of pea, rice alpha-amylase; -10 to -30 in the case of animal genes. Near upstream elements (NUE) in Arabidopsis. Poly A signal found in rice alphaamylase; -10 to -30 in the case of animal genes. Consensus sequence for plant polyadenylation signal. Brassica napus +521, -171, -663 One of two co-dependent regulatory elements responsible for pollen specific activation of tomato lat52 gene; Found at -72 to -68 region; Also found in the promoter of tomato endo-beta-mannanase gene. Lycopersicon esculentum SCGAYNRNNN NNNNNNNNNN NNHD + 99 Chlamydomon as reinhardtii QELEMENTZM ZM13 AGGTCA + 616 Consensus sequence of PRE (plastid response element) in the promoters of HSP70A in Chlamydomonas; Involved in induction of HSP70A gene by both MgProto and light. "Q (quantitative)-element" in maize ZM13 gene (pollen-specific maize gene.) promoter; Found at -107 to 102; Involved in expression enhancing activity. RAV1AAT REALPHALGL HCB21 CAACA AACCAA -184, -336 -407 Lemna gibba ROOTMOTIFTA POX1 S1FBOXSORPS 1L21 ATATT -395, -582 ATGGTA -19 "REalpha" found in Lemna gibba Lhcb21 gene promoter; Located at 134 to -129; Binding site of proteins of whole-cell extracts; The DNA binding activity is high in etiolated plants but much lower in green plants; Required for phytochrome regulation. Motif found both in promoters of rolD "S1F box" conserved both in spinach, RPS1 and RPL21 genes encoding the plastid ribosomal protein S1 and L21, respectively; Negative element; Pisum sativum, Oryza sativa, Arabidopsis thaliana. Oryza sativa, animal Zea mays Zea mays Agrobacteriu m rhizogenes Spinacia oleracea SEF4MOTIFGM 7S RTTTTTR -47 TAAAGSTKST1 TAAAG -16 TATABOXOSP AL TATTTAA +206 TATAPVTRNA LEU TTTATATA +457 TATCCACHVA L21 TATCCAC + 650 TATCCAOSAM Y TATCCA +643, + 650 TATCCAYMOT IFOSRAMY3D TGACGTVMA MY TATCCAY +650 TGACGT + 224 WBBOXPCWR TTTGACY + 604, -497 Might play a role in downregulating RPS1 and RPL21 promoter activity. Consensus sequence found in 5'upstream region (-199) of betaconglycinin (7S globulin) gene; "Binding with SEF4 (soybean embryo factor 4. Target site for trans-acting StDof1 protein controlling guard cellspecific gene expression; KST1 gene encodes a K+ influx channel of guard cells. Binding site for OsTBP2, found in the promoter of rice pal gene encoding phenylalanine ammonialyase; OsTFIIB stimulated the DNA binding and bending activities of OsTBP2 and synergistically enhanced OsTBP2-mediated transcription from the pal promoter. Frequently observed upstream of plant tRNA genes; Found in maize glycolytic glyceraldehyde-3-phospate dehydrogenase 4 (GapC4) gene promoter; Binding site of TATA binding protein (TBP). "TATCCAC box" is a part of the conserved Cis-acting response complex (GARC) that most often contain three sequence motifs, the TAACAAA box , GA-responsive element (GARE); the pyrimidine box, CCTTTT ; and the TATCCAC box, which are necessary for a full GA response "TATCCA" element found in alphaamylase promoters of rice at positions 90 to 150bp upstream of the transcription start sites; Binding sites of OsMYBS1, OsMYBS2 and OsMYBS3 which mediate sugar and hormone regulation of alpha-amylase gene expression. TATCCAY motif and G motif are responsible for sugar repression. Required for high level expression of alpha-Amylase in the cotyledons of the germinated seeds "WB box"; WRKY proteins bind Glycine max Solanum tuberosum Oryza sativa Phaseolus vulgaris, Zea mays Hordeum vulgare Oryza sativa Oryza sativa Vigna mungo Ipomoea KY1 specifically to the DNA sequence motif (T)(T)TGAC(C/T), which is known as the W box WBOXATNPR1 TTGAC WBOXHVISO1 TGACT +223, +605, -498, -570, -618 +143, -476, -497 WBOXNTERF3 TGACY +143, +606, -476, -497, -617 WRKY71OS TGAC +143, +224, +606, -477, -498, -570, -618 WUSATAg +556, -327 "W-box" found in promoter of Arabidopsis thaliana NPR1 gene; SUSIBA2 bind to W-box element in barley iso1 (encoding isoamylase1) promoter "W box" found in the promoter region of a transcriptional repressor ERF3 gene in tobacco; "A core of TGAC-containing Wbox" of, e.g., Amy32b promoter; Binding site of rice WRKY71, a transcriptional repressor of the gibberellin signaling pathway. Target sequence of WUS in the intron of AGAMOUS gene in Arabidopsis (+) stands for 5'-3' orientation and (-) stands for 3'-5' orientation batatas, Triticum aestivum, Hordeum vulgare, Avena fatua, Petroselium crispum, Arabidopsis thaliana Arabidopsis thaliana Hordeum vulgare Nicotiana tabacum Oryza sativa, Petroselinum crispum Oryza sativa