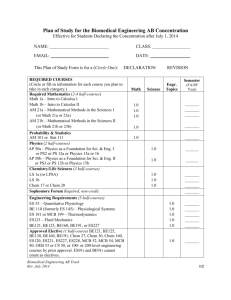
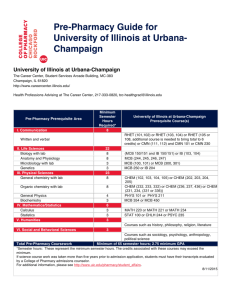
lecture
advertisement

Question What is the chemical nature of the repressor? 1 MCB 140 – 11/8/2006 If you think about it … • • The repressor has to directly bind to a specific DNA sequence (the operator) in the E. coli genome. From a reverse-engineering perspective, the simplest way to design something that interacts with DNA sequencespecifically is to use a nucleic acid that is complementary to that DNA! 2 MCB 140 – 11/8/2006 • In 1959, a biochemical experiment was done “proving” that the lac repressor is not a protein, and is most likely an RNA molecule • In 1965, a genetic experiment was done proving that biochemical experiment entirely wrong 3 MCB 140 – 11/8/2006 1959: (When the z+ gene and the i+ gene arrive in an i- cytoplasm (no repressor), the z+ gene becomes active, and stays active for about 2 hours. At that point, the repressor is made, and shuts the z+ gene off. This offers an elegant opportunity to determine the biochemical nature of the repressor: add an inhibitor of protein synthesis, and let the cells spend their first two hours post-mating in that good stuff. A.B. Pardee and L.S. Prestidge BBA 36: 545. If the repressor is a protein, then inhibiting its synthesis following transfer of z+ into an i- cytoplasm should allow for constitutive synthesis of b-galactosidase! 4 MCB 140 – 11/8/2006 The result • Inhibiting protein synthesis does not destroy the “plateau” effect: b-gal still goes off! • Conclusion: “… the repressor probably is not a protein, since it was made when [protein] synthesis was inhibited. Ribonucleic acid would seem a likely candidate for the role of the repressor.” A.B. Pardee and L.S. Prestidge BBA 36: 545. 5 MCB 140 – 11/8/2006 A distinction between genetics and biochemistry • The validity of Pardee’s conclusion is unequivocally dependent on a biochemical phenomenon (total inhibition of protein synthesis by 5-methyltryptophan) • It turns out that not all protein synthesis in E. coli is inhibited by 5-me-T… • A genetic experiment (=crosses between strains of different genotype) was performed to prove Pardee wrong 6 MCB 140 – 11/8/2006 8.28 7 MCB 140 – 11/8/2006 Let’s mate 1. Make an i- strain of E. coli (constitutive) 2. Make several such strains (=different kinds of mutations) 3. Mate those i- E. coli to other E. coli carrying nonsense suppressor tRNA genes 4. See what happens. 8 MCB 140 – 11/8/2006 “Suppression of and complementation among mutants of the regulatory gene of the lactose operon of Escherichia coli.” Bourgeois S, Cohn M, Orgel LE. JMB 14: 300 (1965) Cross an i- strain with a tRNAsup strain Measure activity of b-gal (+ or - lactose in the medium). 1. 2. Suppressor: lactose Strain i+ i- none (wt) no yes 8 4400 su1 no 6 yes 4400 su3 no 14 yes 3900 7600 7000 1200 1550 3400 7100 9 MCB 140 – 11/8/2006 In their own words “We know … that the suppressor strains used here act at the level of translation by allowing the chain-terminating codon to be read as an amino acid. We have shown suppression of i- mutations by these suppressors and conclude therefore that the i gene of the lactose operon codes for a protein.” 10 MCB 140 – 11/8/2006 stimulus + + Regulation of genes occurs via the interaction of transacting factors (proteins) with cis-acting sequences near the genes themselves. 11 MCB 140 – 11/8/2006 François Jacob: “If it’s true for E. coli, it must be true for E. lephant.” 12 MCB 140 – 11/8/2006 Budding (brewer’s and baker’s) yeast, Saccharomyces cerevisiae 13 MCB 140 – 11/8/2006 Yeast ferment all available sugar even in the presence of oxygen Mammals: sugar glycolysis respiration pyruvate TCA+O.P. CO2+H2O S.c.: sugar glycolysis pyruvate fermentation C2H5OH+CO2 14 MCB 140 – 11/8/2006 A major evolutionary conservation of cellular response to sugar in the medium In Saccharomyces cerevisiae (budding yeast = a fungus = a eukaryote): 1. Enzymes for utilization of the sugar galactose are induced ~1000-fold by galactose. 2. These enzymes are also severely repressed by glucose in the medium. 3. Thus, for these genes to be induced fully, the medium must contain galactose and no glucose. Just like E. coli. 15 MCB 140 – 11/8/2006 Analogy and homology as tools in genetic investigation Animal Mandibular Arch (ventral) Mandibular Arch (dorsal) Hyoid Arch (dorsal) Shark Meckel's cartilage Palatoquadrate cartilage Hyomandibular cartiliage Amphibian Articular (bone) Quadrate (bone) Stapes Mammal Malleus Incus Stapes 16 MCB 140 – 11/8/2006 17 MCB 140 – 11/8/2006 First experiment (Howard Douglas, 1963) Goal: make i- yeast – that is, yeast that synthesize galactose-metabolizing enzymes constitutively. 1. Take mutant strain (gal3) that has a markedly delayed response to galactose and does not grow on it very well at all. 2. Grow on galactose – see what grows. Douglas HC, Penroy G (1963) A gene controlling inducibility of the galactose pathway enzymes in Saccharomyces. Biochim Biophys Acta 68: 155. 18 MCB 140 – 11/8/2006 What grew 1. One would expect revertants of the gal3 mutation. Those didn’t show up. 2. What did show up was true i- cells – yeast that synthesized the GAL enzymes constitutively (that’s why they grew). 3. They made i+ / i- cells – they were inducible. 19 MCB 140 – 11/8/2006 Conclusion (ta-daaa!) “The inducibility in gene in yeast fits the description proposed by Monod and Jacob for regulator genes. … By analogy with the lactose system in E. coli, the galactoseinducible state in yeast corresponds to the production of a repressor, due to i+, while the constitutive state, due to i-, represents a failure to repression.” 20 MCB 140 – 11/8/2006 Nothin’ but net Indeed, the i+ gene is now called GAL80. Its product, Gal80p, is a repressor of GAL genes. 21 MCB 140 – 11/8/2006 But “We might have expected, by further analogy with the systems in E. coli, to find mutations linked to the structural galactose genes and to be expressed as cis dominant constitutives, but these have not yet been detected in our material.” In other words, they wanted to find “operator” mutations, and didn’t find them. 22 MCB 140 – 11/8/2006 Screen for gal cells H. Douglas, D. Hawthorne (1964): 1. Take wt haploid yeast. 2. Zap them with UV light. 3. Replica-plate to find those that are gal. 4. 1:1000 are mutant. Douglas HC, Hawthorne D (1964) Enzymatic expression and genetic linkage of genes controlling lactose utilization in Saccharomyces. Genetics 49: 837. 23 MCB 140 – 11/8/2006 7.5a 24 MCB 140 – 11/8/2006 A.8 25 MCB 140 – 11/8/2006 Initial analysis of mutants 1. 2. 3. 4. Cross mutant to wt. Sporulate. Dissect the tetrad (ascus) Confirm that 2 spores are GAL and 2 are gal. 26 MCB 140 – 11/8/2006 What came out Large number of mutants in different genes: • GAL1 six mutants • GAL2 five mutants • GAL3 six mutants • GAL4 two mutants • GAL5 nine mutants • GAL7 four mutants • GAL10 one mutant 27 MCB 140 – 11/8/2006 “When genes are linked, PDs exceed NPDs” 5.18 28 MCB 140 – 11/8/2006 Linkage analysis Gene pair gal1/gal7 gal1/gal10 gal7/gal10 gal1/gal4 gal3/gal4 PD 313 59 72 21 20 # of tetrads NPD 0 0 0 23 13 T 0 0 0 56 48 29 MCB 140 – 11/8/2006 Conclusions • The following genes are very closely linked: GAL1, GAL7, and GAL10 • The GAL4 gene is not linked to those three 30 MCB 140 – 11/8/2006 What does what Genotype wild type gal1 gal7 gal10 gal10/igal1/gal7 gal1/gal10 gal4 gal4/i- Kinase 13-24 0 6 2.7 13 0 0 0 0 Enzymatic activities Transferase Epimerase 9-14 8-34 14.9 87.2 0 33.8 2.2 0 15.5 0 0 20.3 6.1 0 0 0 0 0 31 MCB 140 – 11/8/2006 The fun part of this complete breakfast The gal4 mutation is unlinked to the enzyme genes, and yet the GAL4 gene product is required for their synthesis. 32 MCB 140 – 11/8/2006 Ta-daaa!! “The closely linked genes GAL1, GAL7, and GAL10 [code for] the galactose pathway enzymes, galactokinase, transferase, and epimerase. The mutation gal4 blocks the synthesis of these enzymes, but unlike the phenotypically similar mutation Oo in E. coli is complementable and is not linked to the genes whose expression it controls.” 33 MCB 140 – 11/8/2006 Summary ctd. “GAL4 apparently produces a cytoplasmic product required for the expression of GAL1, GAL7, and GAL10. The role of the repressor might be to prevent the synthesis or the activity of this product.” 34 MCB 140 – 11/8/2006 But still “In considering a problem of an operator locus in yeast, it is of interest that [we] have been unable to find a constitutive mutant analogous to an OC type…” = cis dominant mutation that leads to constitutive galactose enzyme synthesis “find mutation in operator that does not bind repressor…” 35 MCB 140 – 11/8/2006 1966: got it! Complex mutagenesis screen in a diploid strain for constitutive mutants – exactly analogous to such screen in E. coli, where a diploid was used to prevent isolation of mutants in the repressor gene itself. Got one dominant mutation that was very tightly linked to the GAL4 gene. Douglas HC, Hawthorne D (1966) Regulation of genes controlling synthesis of the galactose pathway enzymes in yeast. Genetics 54: 911. 36 MCB 140 – 11/8/2006 - galactose Gal80p GAL7 GAL4 GAL10 GAL1 37 MCB 140 – 11/8/2006 + galactose Gal80p GAL4 Gal4p GAL7 GAL10 GAL1 38 MCB 140 – 11/8/2006 - galactose in gal81 strain Gal80p gal81 GAL4 Gal4p GAL7 GAL10 GAL1 39 MCB 140 – 11/8/2006 wrong (correct between 1966-1978) 40 MCB 140 – 11/8/2006 Gal4p is synthesized at all times, irrespective of the presence of galactose Yasuji Oshima (Osaka) 41 MCB 140 – 11/8/2006 Tokyo Kokuritsu Hakubutsukan (Ueno) 42 MCB 140 – 11/8/2006 sumi-e 43 MCB 140 – 11/8/2006 gal4-4 – a ts allele of GAL4 GAL at 25 and gal at 35 1. Take wt and gal4-4 cells. 2. Grow at 35 in glucose 3. Move them to 25 4. Immediately add galactose 5. Measure galactokinase. 6. Compare wt and gal4-4. gal4-4 (3525) gal4-4 (25) wild-type 44 Matsumoto et al. J. Bacteriol. 134: 446 (1978). MCB 140 – 11/8/2006 Epistasis (1982-84) A gal80 cell synthesizes galactose utilization enzymes constitutively. A gal4 cell does not synthesize those enzymes under any condition. A double-mutant gal80 gal4 cell has the same phenotype as a gal4 cell – no enzyme synthesis. GAL4 is epistatic to GAL80. Gal4p acts downstream of Gal80p. A “superrepressor” allelic form of GAL80 (GAL80S) does not respond to galactose. Overexpression of Gal4p can overcome its effect! That is, GAL4 GAL80S is uninducible, but GAL4high copy GAL80S is inducible. Johnston and Hopper PNAS 79: 6971 (1982). Torchia et al. MCB 4: 1521 (1984). 45 MCB 140 – 11/8/2006 SUPPRESSION: A given mutation (A) has a discrete phenotype that is not normal, i.e. not wild-type. The presence of the second mutation (B, the suppressor mutation) causes the AB double mutant to display a phenotype that is normal or near-normal. Thus, a suppressor mutation rescues or restores or repairs, in whole or in part, the defect caused by the first mutation. Examples: A nonsense mutation in a gene can be suppressed by a mutation in a tRNA gene in which the anticodon has been mutated to read the nonsense codon (informational suppressor). A temperature-sensitive mutation that destabilizes a protein can be corrected by a compensating mutation in another gene product that binds to and acts in a complex with the first gene product (extragenic suppressor). Overproduction of a transcription factor can overcome the need for a protein kinase in the pathway that is normally needed to activate that transcription factor (dosage suppressor). Loss of a repressor of a gene can compensate for absence of the positive regulator normally needed to turn on that gene (bypass suppressor). EPISTASIS: Two mutations (A and B) each have discrete phenotypes that are readily distinguishable from each other, and neither are normal (both are non-wild-type). If the phenotype of the AB double mutant resembles that of one of the two single mutants and not the other, that mutation is said to be epistatic over the other. Thus, if the AB double mutant looks like the A mutant alone, mutation A is said to be epistatic over mutation B; conversely, if the AB double mutant looks like the B mutant alone, mutation B is said to be epistatic over mutation A. Examples: In yeast, the ade3 mutation blocks purine biosynthesis early in the pathway, whereas the ade2 mutation blocks the pathway later. Both require Ade in the medium to grow. The ade2 mutant has a white colony color, but the ade2 mutant has a pinkto-red colony color (because the metabolic intermediate that accumulates in the ade2 mutant polymerizes to form a pigment). The ade2 ade3 double mutant still requires Ade to grow, but displays a white colony color. Thus, the ade3 mutation is said to be epistatic to the ade2 mutation (which make sense because ade3 blocks production of the intermediate that would otherwise accumulate in the ade2 mutant). In flies, apterous (ap) mutations block wing formation, whereas curled wing (cw) causes a dramatic change in wing morphology. Neither have normal wings. A ap cw double mutant has a wingless phenotype. Thus, ap is epistatic to cw. Thus, in the pathway for construction of a wing, ap functions before cw. Prof. Jeremy Thorner, UCB 46 MCB 140 – 11/8/2006 Also “described” on pp. 592-593, under the amusing heading “The yeast GAL system is another complex regulatory mechanism.” 47 MCB 140 – 11/8/2006 How genes respond to environmental stimuli 48 MCB 140 – 11/8/2006 More from Dr. Jacob “I have always been convinced that the same principles operating in bacteria are also operating in higher organisms with added complexity. The question therefore is to understand what kind of complexity is involved and how it is generated.” 49 MCB 140 – 11/8/2006 Next time The answer to Dr. Jacob’s question. 50 MCB 140 – 11/8/2006 Further reading 1. T. Brock The Emergence of Bacterial Genetics (CSHL Press 1990) 2. M. Ptashne, A. Gann Genes and Signals (ibid) 3. H. Judson The eighth day of creation (Simon and Schuster 1979) 4. S. Brenner My Life in Science (BMC Press 2001) 51 MCB 140 – 11/8/2006