Powerpoint slides
advertisement

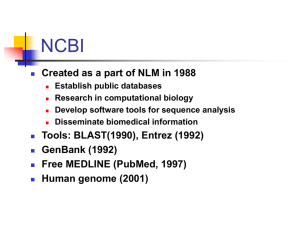
A Field Guide to GenBank and NCBI Molecular Biology Resources slightly modified from Peter Cooper ftp://ftp.ncbi.nih.gov/pub/cooper/FieldGuide/ Eric Sayers ftp://ftp.ncbi.nih.gov/pub/sayers/Field_Guide/U_Penn/ NCBI Resources • About NCBI • NCBI Sequence Databases – Primary Database – GenBank – Derivative Databases - RefSeq • Entrez Databases and Text Searching • BLAST Services • Genomic Resources The National Center for Biotechnology Information (NCBI) • Created as a part of NLM in 1988 – – – – • • • • Establish public databases Perform research in computational biology Develop software tools for sequence analysis Disseminate biomedical information Tools: BLAST(1990), Entrez (1992) GenBank (1992) Free MEDLINE (PubMed, 1997) Human genome (2001) NCBI Home Page http://www.ncbi.nlm.nih.gov To learn more, visit the “Site Map” and “About NCBI” web pages About NCBI Some NCBI Statistics…. 23 22 21 20 19 18 17 16 15 14 13 12 11 10 9 8 7 6 5 4 3 2 1 0 1982 30000 28000 26000 24000 22000 20000 18000 16000 14000 12000 10000 Base Pairs Sequences 8000 6000 4000 2000 0 1984 1986 1988 1990 1992 1994 1996 1998 2000 2002 Base Pairs of DNA (millions) Sequences (millions) Growth of GenBank Users per day 250000 1997 1998 1999 2000 200000 150000 100000 50000 Christmas Day 0 2001 Molecular Databases • Primary Databases – Original submissions by experimentalists – Database staff organize but don’t add additional information • Example: GenBank • Derivative Databases – Human curated • compilation and correction of data • Example: SWISS-PROT, NCBI RefSeq mRNA – Computationally Derived • Example: UniGene – Combinations • Example: NCBI Genome Assembly What is GenBank? NCBI’s Primary Sequence Database • Nucleotide only sequence database • GenBank Data – Direct submissions individual records (BankIt, Sequin) – Batch submissions via email (EST, GSS, STS) – ftp accounts established for sequencing centers • Data shared amongst three collaborating databases: – GenBank – DNA Database of Japan (DDBJ). – European Molecular Biology Laboratory Database (EMBL) The International Nucleotide Sequence Database Collaboration NIH Sequin BankIt ftp Entrez NCBI GenBank •Submissions •Updates •Submissions •Updates EMBL CIB NIG DDBJ •Submissions •Updates getentry EBI SRS EMBL GenBank: NCBI’s Primary Sequence Database Release 133 22,318,883 28,507,990,166 110,000 + December 2002 Records Nucleotides Species • full release every two months • incremental and cumulative updates daily • available only through internet ftp://ftp.ncbi.nih.gov/genbank/ >90 Gigabytes of data Entrez Nucleotide RefSeq 1% EMBL 9% DDBJ 19% GenBank 71% 23,464,770 records Primary vs. Derivative Databases Curators RefSeq Sequencing Centers TATAGCCG AGCTCCGATA CCGATGACAA Labs Genome Assembly TATAGCCG TATAGCCG TATAGCCG TATAGCCG GenBank UniGene Algorithms Traditional GenBank Divisions •Direct Submissions (Sequin and BankIt) •Accurate •Well characterized BCT INV MAM PHG PLN PRI ROD SYN VRL VRT Bacterial and Archeal Invertebrate Mammalian (ex. ROD and PRI) Phage Plant and Fungal Primate Rodent Synthetic (cloning vectors) Viral Other Vertebrate A Traditional GenBank Record Locus Field Molecule Type Definition Line Accession Number Version GI (GenInfo) Keywords Taxonomy Modification Date GenBank Division A Traditional GenBank Record Bulk Sequence Divisions of GenBank •Batch Submissions (email and ftp) •Inaccurate •Poorly Characterized EST STS GSS HTG HTC Expressed Sequence Tag Sequence Tagged Site Genome Survey Sequence High Throughput Genomic High Throughput cDNA Organization of GenBank 11 Traditional Divisions Traditional 8% PAT 4% 1 Patent Division STS, HTG, HTC 2% GSS 19% EST 67% 5 Bulk Divisions 23,087,196 records What is UniGene? A gene-oriented view of sequence entries •MegaBlast-based automated sequence clustering •Nonredundant set of gene-oriented clusters •Each cluster represents a unique gene •Provides information on tissue-specific expression and map locations •Includes well-characterized genes and novel ESTs •Useful for gene discovery and selection of mapping reagents Organisms Represented in UniGene Genome Sequencing Whole BAC insert (or genome) shredding sequencing GSS division or trace archive cloning isolating assembly Draft Sequence (HTG division) Working Draft Sequence gaps HTG Division: High Throughput Genome phase 1 HTG phase 2 HTG phase 3 ROD Acc = AC109609.1 Acc =AC109609.6 Acc = AC109609.10 HTG Division: High Throughput Genome NCBI’s Third Party Annotation (TPA) Database NEW • NCBI now accepts the submission of new annotations of existing GenBank sequences; • Facilitates the annotation of genomes by experts; A Sample TPA record RefSeq: NCBI’s Derivative Sequence Database • Curated transcripts and proteins – reviewed – human, mouse, rat, fruit fly, zebrafish, arabidopsis • Human model transcripts and proteins • Assembled Genomic Regions (contigs) – draft human genome – mouse genome • Chromosome records – Microbial – viral – organelle The RefSeq Accession Numbers mRNAs and Proteins NM_123456 NP_123456 NR_123456 XM_123456 XP_123456 XR_123456 Gene Records NG_ 123456 Assemblies NT_ 123456 NW_123456 NC_ 123456 NR_ 123456 human Curated mRNA mouse rat Curated Protein fruit fly Curated non-coding RNA zebrafish Predicted Transcript (human, mouse) Arabidopsis Predicted Protein (human, mouse) Predicted non-coding RNA Reference Genomic Sequence (human) Contig (Mouse and Human) Supercontig (Mouse) Chromosome (Microbial,Viral,Arabidopsis ) Interim Identifier for Microbial Chromosomes Curated RefSeq Records: NM_, NP_ Entrez: Linking and Neighboring The Entrez Databases The (ever) Journals Expanding Entrez System UniGene Books PubMed Central SNP PubMed UniSTS Nucleotide Protein PopSet ProbeSet Entrez Genome Structure Taxonomy CDD 3D Domains OMIM Entrez Nucleotides glucose 6 phosphate dehydrogenase Document Summaries: glucose 6 phosphate dehydrogenase[All Fields] = 748 hits Entrez Nucleotides: Limits Accession All Fields Author Name EC/RN Number glucose 6 phosphate dehydrogenase Feature key Filter Gene Name Issue Journal Name Keyword Modification Date Organism Page Number Primary Accession Properties Protein Name Publication Date SeqID String Sequence Length Substance Name Text Word Entrez Nucleotides: Preview/Index Adding Terms: Preview/Index Accession All Fields Author Name EC/RN Number Feature key Filter Gene Name Issue Journal Name Keyword Modification Date Organism Page Number Primary Accession Properties Protein Name Publication Date SeqID String Sequence Length . . . Plant G6PD mRNAs Display: Formats, Links, and Neighbors Summary Brief ASN.1 FASTA XML GenBank GI list LinkOut Nucleotide Neighbors Genome Links ProbeSet Links OMIM Links PopSet Links Protein Links PubMed Links SNP Links Structure Links Taxonomy Links >gi|603218|gb|U18238.1|MSU18238 Medicago sativa glucose-6-phosphate dehyd CCACCAGATATAATTAAGTAGATCAGAGTAGAAGAAGATGGGAACAAATGAATGGCATGTAGAAAGAAGA GATAGCATAGGTACTGAATCTCCTGTAGCAAGAGAGGTACTTGAAACTGGCACACTCTCTATTGTTGTGC TTGGTGCTTCTGGTGATCTTGCCAAGAAGAAGACTTTTCCTGCACTTTTTCACTTATATAAACAGGAATT GTTGCCACCTGATGAAGTTCACATTTTTGGCTATGCAAGGTCAAAGATCTCCGATGATGAATTGAGAAAC AAATTGCGTAGCTATCTTGTTCCAGAGAAAGGTGCTTCTCCTAAACAGTTAGATGATGTATCAAAGTTTT TACAATTGGTTAAATATGTAAGTGGCCCTTATGATTCTGAAGATGGATTTCGCTTGTTGGATAAAGAGAT TTCAGAGCATGAATATTTGAAAAATAGTAAAGAGGGTTCATCTCGGAGGCTTTTCTATCTTGCACTTCCT > CCTTCAGTGTATCCATCCGTTTGCAAGATGATCAAAACTTGTTGCATGAATAAATCTGATCTTGGTGGAT GGACACGCGTTGTTGTTGAGAAACCCTTTGGTAGGGATCTAGAATCTGCAGAAGAACTCAGTACTCAGAT gi number TGGAGAGTTATTTGAAGAACCACAGATTTATCGTATTGATCACTATTTAGGAAAGGAACTAGTGCAAAAC Locus name ATGTTAGTACTTCGTTTTGCAAATCGGTTCTTCTTGCCTCTGTGGAACCACAACCACATTGACAATGTGC AGATAGTATTTAGAGAGGATTTTGGAACTGATGGTCGTGGTGGATATTTTGACCAATATGGAATTATCCG Database identifiers AGATATCATTCCAAACCATCTGTTGCAGGTTCTTTGCTTGATTGCTATGGAAAAACCCGTTTCTCTCAAG Accession number gb GenBank CCTGAGCACATTCGAGATGAGAAAGTGAAGGTTCTTGAATCAGTACTCCCTATTAGAGATGATGAAGTTG TTCTTGGACAATATGAAGGCTATACAGATGACCCAACTGTACCGGACGATTCAAACACCCCGACTTTTGC emb EMBL AACTACTATTCTGCGGATACACAATGAAAGATGGGAAGGTGTTCCTTTCATTGTGAAAGCAGGGAAGGCC dbj DDBJ CTAAATTCTAGGAAGGCAGAGATTCGGGTTCAATTCAAGGATGTTCCTGGTGACATTTTCAGGAGTAAAA AGCAAGGGAGAAACGAGTTTGTTATCCGCCTACAACCTTCAGAAGCTATTTACATGAAGCTTACGGTCAA sp SWISS-PROT GCAACCTGGACTGGAAATGTCTGCAGTTCAAAGTGAACTAGACTTGTCATATGGGCAACGATATCAAGGG pdb Protein Databank ATAACCATTCCAGAGGCTTATGAGCGTCTAATTCTCGACACAATTAGAGGTGATCAACAACATTTTGTTC GCAGAGACGAATTAAAGGCATCATGGCAAATATTCACACCACTTTTACACAAAATTGATAGAGGGGAGTT pir PIR GAAGCCGGTTCCTTACAACCCGGGAAGTAGAGGTCCTGCAGAAGCAGATGAGTTATTAGAAAAAGCTGGA prf PRF TATGTTCAAACACCCGGTTATATATGGATTCCTCCTACCTTATAGAGTGACCAAATTTCATAATAAAACA ref RefSeq AGGATTAGGATTATCAGGAGCTTATAAATAAGTCTTCAATAAGCTTGTGAAATTTTCGTTATAATCTCTC TCATTTTGGGGTGTATATCAAGCATTTAAGCGCGTGTTTGACACAGTTTGTGTAATAGATTTGGCTCTGA ATGAAAATAAACGGGAATTGTTTCTTTTTGTTTTA FASTA definition line >gi|603218|gb|U18238.1|MSU18238 Entrez Genome Organism Pages The Map Viewer: a common platform for integrated display The Map Viewer Entrez PubMed Online Books Entrez Specialized Databases Taxonomy Searchable taxonomic tree having nodes for all species with records in an Entrez database OMIM Online Mendelian Inheritance in Man: A database of genetically linked human diseases ProbeSet Expression data (GEO) and microarray datasets Entrez Taxonomy Entrez OMIM Entrez ProbeSet Trace Archive Entrez Structure Structure Summary Cn3D viewer Related Structures Conserved Domains Cn3D: Displaying Structures Structural Alignment