What is a Database
advertisement

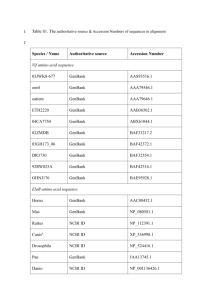
Database Resources The National Center for Biotechnology Information (NCBI) a primary resource for molecular biology information www.ncbi.nih.gov NCBI Mission …. to develop new information technologies to aid in the understanding of fundamental molecular and genetic processes that control health and disease. What does this involve ? • creating automated systems for storing and analyzing knowledge about molecular biology, biochemistry, and genetics; • facilitating the use of such databases and software by the research and medical community; • coordinating efforts to gather biotechnology information both nationally and internationally; • performing research into advanced methods of computer-based information processing for analyzing the structure and function of biologically important molecules. What is a Database ? • A model or representation of some aspect of the real world • An organized collection of data. May contain many different types of data • Coherent, consistent and designed for a specific purpose • A computational system for managing and querying the data. What is a Database ? • A collection of information organized in such a way that a computer program can quickly select desired pieces of data. • An electronic filing system • Traditional databases are organized by fields, records, and files. • A field is a single piece of information; • a record is one complete set of fields • a file is a collection of records. For example, a telephone book is analogous to a file. It contains a list of records, each of which consists of three fields: name, address, and telephone number. What is a Database ? • To access information from a database, you need a database management system (DBMS). This is a collection of programs that enables you to enter, organize, and select data in a database. • Most molecular biology databases primarily use relational database management systems (RDBMS). Relational Database • A relational database is like a large spreadsheet. Each field is a column, each row is an entry. Relational databases use a set of tables to organize data. • Each entry must be unambiguously identified • Names are not reliable e.g. incorrectly assigned gene function • Unique IDs (UID)s are used, e.g. in GenBank these are called accession numbers UID Name Sequence Quality Value BU039022 PP_LEa0001A01f CATACAAAT … 35 BU039057 PP_LEa0001B17f TACGGCTAC … 28 Relational Database • Achieving consistency • Repeated information is stored in a single place. • Only one copy needs to be updated Sequence UID Definition Locus Accession Taxonomy ID* Sequence Taxonomy Taxonomy ID* Genus Species Ref Index* UID Medline ID * May be referred Ref Index Medline ID Authors Title Journal * May be referred to indirectly via an index to by a secondary ID Relational Database • Language used is SQL or structured query language • Easy to understand (essentially English?) • Relatively consistent across RBDMS • Supplies a set of commands to define tables, insert data and make queries • Queries • SELECT some fields FROM some table WHERE some condition is met • E.g. select accession, sequence FROM sequence WHERE Accession = BU039022 BU039022 CATACAAATACTGCTACHTAAATC …. • More complex queries require two or more tables be joined to produce a result Relational Database • Most RDBMS do not allow users to directly query the database by SQL. • An ill formed query can overload or crash the system • SQL still too complex for biologists? • Provide a search interface for the user instead • E.g. user enters a phrase and the database identifies what part of the database should be searched. • The queries that make it through the web interface have to be translated to SQL Relational database : Example GenBank Query What Constitutes a Good Database ? • • • • • • Broad coverage of the chosen topic Up to date information gathering Curated Support staff Commitment to the future Good query interface Issues for Molecular Biological Databases ? • • • • Annotation Archives Updates Redundancy Issues for Molecular Biological Databases ? • Annotation • Adding biological information to genome sequence. Textual descriptive information • Correctness • Many genes are incorrectly annotated. May assign a function to a novel gene from a similar sequence that may itself be incorrectly annotated so the error is propagated throughout the database. • Routine error • Quality • Expert or non expert curation? Who provided the curation? • Is there any biological verification? • What vocabulary is used • Has their been any peer review ? Issues for Molecular Biological Databases ? • Archival Quality • • Is the database archival or curated Can the same data be recovered later • Don’t overwrite primary key (each accession numbers) • The best databases note any changes to the data. • Updates • • • • How often is the database updated? Major databases take direct submissions • Only the direct submitter can make changes, even if you can prove its wrong. When is a sequence finished ? How is annotation updated as more knowledge is available • Redundancy • • This is a major issue, how do we deal with it without losing potentially valuable information. Also relates to archival quality NCBI and GenBank • Genbank is the genetic sequence database of all publicly available DNA and derived protein sequences, with annotations describing the biological information in them. • GenBank is hosted within NCBI • Researchers submit their sequences to GenBank • NCBI provides analysis and retrieval resources for the data in GenBank (and many other NCBI hosted databases). NCBI Databases (http://www.ncbi.nlm.nih.gov/guide/all/#Databases_) • • • • • • • • • • • • • Nucleotide Database EST (dbEST) GSS (dbGSS) Protein Database Structure Database Genome 3D Domains Conserved Domains UniSTS Gene UniGene HomoloGene Reference Sequence (refseq) • • • • • • • • • • • • • • SNP (dbSNP) dbVAR – large scale genomic variation dbGAP – integration of genotype & phenotype PopSet Database Taxonomy Database GEO Profiles GEO Datasets Cancer Chromosomes Epigenomics PubMed Central Journals MeSH Bookshelf OMIM Database Retrieving Data from NCBI using Entrez • Entrez is a text based retrieval system that integrates all the information resources available at the NCBI such as; 1. 2. 3. 4. 5. 6. 7. Scientific literature DNA and protein sequence databases 3D protein structure and protein domain data Population study datasets Expression data Assemblies of complete genomes Taxonomic information http://www.ncbi.nlm.nih.gov/guide/all/#howto_ Create/login to the myNCBI portal Understanding GenBank records Go to http://www.ncbi.nlm.nih.gov/Sitemap/samplerecord.html#ModificationsDateB Click on the links on the left to get a description of what the term means, Copy the description into a word document and after completed, save the document on your drupal web site Entrez Sequences Help http://www.ncbi.nlm.nih.gov/books/NBK44864/