ppt - University of Illinois at Urbana
advertisement

Overview of Biological Databases
(Lecture for CS498-CXZ Algorithms in Bioinformatics)
Sept. 6, 2005
ChengXiang Zhai
Department of Computer Science
University of Illinois, Urbana-Champaign
Most slides are taken from NCBI field guide at the web site http://www.ncbi.nlm.nih.gov/
The Central Dogma & Biological Data
Original DNA Sequences
(Genomes)
Expressed DNA sequences
( = mRNA Sequences
= cDNA sequences)
Expressed Sequence Tags
(ESTs)
Protein Sequences
-Inferred
-Direct sequencing
Protein structures
-Experiments
-Models (homologues)
Literature information
Entrez Integrates Most of Them!
CancerChromosomes
Gene
UniGene
UniST
S
Homologen
e
SNP
Genome
PopSet
Nucleotide
GEO
Books
MeSH
PubMed
OMIM
Entrez
Taxonomy
GEO
Datasets
Protein
PMC
Journal
s
Domains
Structur
e
3D Domains
Outline
• NCBI & Entrez
• Major Biological Databases
• Using Entrez
Some background about
Entrez…
The National Center for
Biotechnology Information
Bethesda,MD
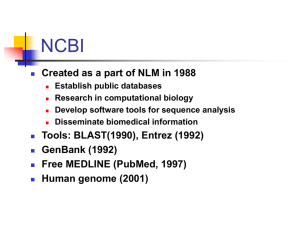
Created in 1988 as a part of the
National Library of Medicine at NIH
– Establish public databases
– Research in computational biology
– Develop software tools for sequence analysis
– Disseminate biomedical information
Web Access: http://www.ncbi.nlm.nih.gov
Number of Users and Hits Per Day
450,000
400,000
1997 1998
1999
2000
2001
2002
2003
Number of Users
350,000
300,000
250,000
Currently averaging
10,000,000 to 50,000,000
hits per day!
200,000
150,000
100,000
50,000
0
Christmas &
New Year’s Days
Major Biological Databases
Entrez: Database Integration
PMC
Cancer Chromosome
OMIM
Word weight
Books
PubMed
Abstracts
Related Articles
PubChem
3D domain
CDD
Phylogeny
Taxonomy
VAST
Gene
HomoloGene
3 -D
Structure
Neighbors
Related Structures
Genome
Genome Project
UniGene
GEO
SNP
OMIM
BLAST
BLAST
Protein
Sequences
Nucleotide
Sequences
Neighbors
Related Sequences
Hard Link
Neighbors
Related Sequences
BLink
Domains
Types of Databases
•
Primary Databases
– Original submissions by experimentalists
– Content controlled by the submitter
• Examples: GenBank, SNP, GEO
•
Derivative Databases
– Built from primary data
– Content controlled by third party (NCBI)
• Examples: Refseq, RefSNP, GEO Datasets,
UniGene, TPA, NCBI Protein, Structure,
Conserved Domain
Primary vs. Derivative
Sequence Databases
RefSeq
Labs
Sequencing
Centers
TATAGCCG
AGCTCCGATA
CCGATGACAA
Curators
TATAGCCG
TATAGCCG
TATAGCCG
TATAGCCG
Updated
continually
by NCBI
GenBank
Updated ONLY
by submitters
Genome
Assembly
UniGene
Algorithms
Entrez Nucleotides
•
•
•
•
Primary
GenBank / EMBL / DDBJ
57,172,944
100%
80%
Derivative
RefSeq
60%
Third
40% Party Annotation
PDB
20%
Total0%
PDB
TPA
1,278,742
RefSeq
4,653
GenBank
5,973
58,462,312
Entrez Protein: Derivative Databases
GenPept
3,515,141
RefSeq
1,802,523
Third
Party Annotation
100%
Swiss
Prot
80%
PIR 60%
PRF 40%
PDB 20%
PDB
4,217
PRF
189,324
PIR
222,232
SwissProt
TPA
12,079
RefSeq
68,621
GenPept
Total0%
5,814,137
BLAST nr total
2,726,372
Database 1: GenBank
NCBI’s Primary Sequence Database
What is GenBank?
•
•
Nucleotide only sequence database
Archival in nature
– Historical
– Reflective of submitter point of view (subjective)
– Redundant
•
GenBank Data
•
– Direct submissions (traditional records)
– Batch submissions (EST, GSS, STS)
– ftp accounts (genome data)
Three collaborating databases
– GenBank
– DNA Database of Japan (DDBJ)
– European Molecular Biology Laboratory (EMBL) Database
International Sequence
Database Collaboration
Entrez
NIH
NCBI
GenBank
•Submissions
•Updates
•Submissions
•Updates
EMBL
CIB
NIG
DDBJ
•Submissions
•Updates
getentry
EBI
SRS
EMBL
GenBank Divisions
“Organismal”
PRI
ROD
PLN
BCT
INV
VRT
VRL
MAM
PHG
SYN
UNA
(28)
(15)
(13)
(11)
(7)
(7)
(4)
(2)
(1)
(1)
(1)
Primate
Rodent
Plant and Fungal
Bacterial/Archeal
Invertebrate
Other Vertebrate
Viral
Mammalian
Phage
Synthetic
Unannotated
EST
GSS
HTG
PAT
STS
CON
(377)
(138)
(63)
(17)
(9)
(1)
Expressed Sequence Tag
Genome Survey Sequence
High Throughput Genomic
Patent
Sequence Tagged Site
Contigs, virtual
• Organized by taxonomy (sort of)
• Direct submissions (Sequin/Bankit)
• Accurate (~1 error per 10,000 bp)
• Well characterized
“Functional”
• Organized by sequence type
• Batch submissions (ftp/email)
• Inaccurate
• Poorly characterized
GenBank Functional (Bulk) Divisions
•
Expressed Sequence Tag
– 1st pass single read cDNA
•
GenBank
EST
GSS
– 1st pass single read gDNA
•
HTG
STS
Genome Survey Sequence
High Throughput Genomic
– incomplete sequences of genomic clones
•
Sequence Tagged Site
– PCR-based mapping reagents
Whole Genome Shotgun
EST Division: Expressed Sequence Tags
>IMAGE:275615 5' mRNA sequence
GACAGCATTCGGGCCGAGATGTCTCGCTCCGTGGCCTTAGCTGTGCTCGCGCTACTCTCTCTTTCTGG
TGGAGGTATCCAGCGTACTCCAAAGATTCAGGTTTACTCACGTCATCCAGCAGAGAATGGAAAGTCAA
TTCCTGAATTGCTATGTGTCTGGGTTTCATCCATCCGACATTGAAGTTGACTTACTGAAGAATGGAGA
5’
GAATTGAAAAAGTGGAGCATTCAGACTTGTCTTTCAGCAAGGACTGGTCTTTCTATCTCTTGTACTAC
30,000
TGAATTCACCCCCACTGAAAAAGATGAGTATGCCTGCCGTGTTGAACCATGTNGACTTTGTCACAGNC
AAGTTNAGTTTAAGTGGGNATCGAGACATGTAAGGCAGGCATCATGGGAGGTTTTGAAGNATGCCGCN
genes
3’
TTGGATTGGGATGAATTCCAAATTTCTGGTTTGCTTGNTTTTTTAATATTGGATATGCTTTTG
nucleus
>IMAGE:275615 3', mRNA sequence
- isolate unique clones
NNTCAAGTTTTATGATTTATTTAACTTGTGGAACAAAAATAAACCAGATTAACCACAACCATGCCTTA
RNA
- sequence once from
TTATCAAATGTATAAGANGTAAATATGAATCTTATATGACAAAATGTTTCATTCATTATAACAAATTT
gene products
each end
AATAATCCTGTCAATNATATTTCTAAATTTTCCCCCAAATTCTAAGCAGAGTATGTAAATTGGAAGTT
CTTATGCACGCTTAACTATCTTAACAAGCTTTGAGTGCAAGAGATTGANGAGTTCAAATCTGACCAAG
GTTGATGTTGGATAAGAGAATTCTCTGCTCCCCACCTCTANGTTGCCAGCCCTC
make cDNA
library
80-100,000 unique
cDNA clones in library
ESTs in Entrez
Total
Human
Mouse
Rat
Zebrafish
Wheat
Barley
Maize
28 million records
6.0 million
4.3 million
0.7 million
0.6 million
0.6 million
0.3 million
0.4 million
GSS, WGS, HTG
Whole BAC insert (or genome)
shred
sequence
GSS division
or trace archive assembly
isolate clones
whole genome shotgun assemblies
(traditional division)
Draft sequence (HTG division)
HTG Example: Honeybee Draft Sequences
LOCUS
AC141845
147720 bp
DNA linear
HTG
19-MAR-2004
DEFINITION Apis mellifera clone CH224-4A2, WORKING DRAFT SEQUENCE,
14 unordered pieces.
ACCESSION AC141845
VERSION
AC141845.1 GI:29124029
KEYWORDS HTG; HTGS_PHASE1; HTGS_DRAFT.
• Unfinished sequences of BACs
• Gaps and unordered pieces
• Finished sequences (Phase 3) move to
traditional GenBank division
50
55
50
40
45
35
40
30
25
Sequence records
Total base pairs
Release 148:
35
45.2 million records
49.4 billion nucleotides
30
25
20
Average doubling time ≈ 14 months
20
15
15
10
10
5
0
5
’83 ’84 ’85 ’86 ’87 ’88 ’89 ’90 ’91 ’92 ’93 ’94 ’95 ’96 ’97 ’98 ’99 ’00 ’01 ’02 ’03 ’04 ’05 ’06
0
Total Base Pairs
(billions)
Sequence Records
(millions)
45
File Formats of the
Sequence Databases
Each sequence is represented by
a text record called a flat file.
GenBank/GenPept (useful for scientists)
FASTA
ASN.1 & XML
(the simplest format)
(useful for programmers)
LOCUS
DEFINITION
AY182241
1931 bp
mRNA
linear
PLN 04-MAY-2004
Malus x domestica (E,E)-alpha-farnesene synthase (AFS1) mRNA,
complete cds.
ACCESSION
AY182241
VERSION
AY182241.2 GI:32265057
KEYWORDS
.
SOURCE
Malus x domestica (cultivated apple)
ORGANISM Malus x domestica
Eukaryota; Viridiplantae; Streptophyta; Embryophyta; Tracheophyta;
Spermatophyta; Magnoliophyta; eudicotyledons; core eudicots;
rosids; eurosids I; Rosales; Rosaceae; Maloideae; Malus.
REFERENCE
1 (bases 1 to 1931)
AUTHORS
Pechous,S.W. and Whitaker,B.D.
TITLE
Cloning and functional expression of an (E,E)-alpha-farnesene
synthase cDNA from peel tissue of apple fruit
JOURNAL
Planta 219, 84-94 (2004)
REFERENCE
2 (bases 1 to 1931)
AUTHORS
Pechous,S.W. and Whitaker,B.D.
TITLE
Direct Submission
JOURNAL
Submitted (18-NOV-2002) PSI-Produce Quality and Safety Lab,
USDA-ARS, 10300 Baltimore Ave. Bldg. 002, Rm. 205, Beltsville, MD
20705, USA
REFERENCE
3 (bases 1 to 1931)
AUTHORS
Pechous,S.W. and Whitaker,B.D.
TITLE
Direct Submission
JOURNAL
Submitted (25-JUN-2003) PSI-Produce Quality and Safety Lab,
USDA-ARS, 10300 Baltimore Ave. Bldg. 002, Rm. 205, Beltsville, MD
20705, USA
REMARK
Sequence update by submitter
COMMENT
On Jun 26, 2003 this sequence version replaced gi:27804758.
FEATURES
Location/Qualifiers
source
1..1931
/organism="Malus x domestica"
/mol_type="mRNA"
/cultivar="'Law Rome'"
/db_xref="taxon:3750"
/tissue_type="peel"
gene
1..1931
/gene="AFS1"
CDS
54..1784
/gene="AFS1"
/note="terpene synthase"
/codon_start=1
/product="(E,E)-alpha-farnesene synthase"
/protein_id="AAO22848.2"
/db_xref="GI:32265058"
/translation="MEFRVHLQADNEQKIFQNQMKPEPEASYLINQRRSANYKPNIWK
NDFLDQSLISKYDGDEYRKLSEKLIEEVKIYISAETMDLVAKLELIDSVRKLGLANLF
EKEIKEALDSIAAIESDNLGTRDDLYGTALHFKILRQHGYKVSQDIFGRFMDEKGTLE
DFLHKNEDLLYNISLIVRLNNDLGTSAAEQERGDSPSSIVCYMREVNASEETARKNIK
GMIDNAWKKVNGKCFTTNQVPFLSSFMNNATNMARVAHSLYKDGDGFGDQEKGPRTHI
LSLLFQPLVN"
ORIGIN
1 ttcttgtatc ccaaacatct cgagcttctt gtacaccaaa ttaggtattc actatggaat
61 tcagagttca cttgcaagct gataatgagc agaaaatttt tcaaaaccag atgaaacccg
121 aacctgaagc ctcttacttg attaatcaaa gacggtctgc aaattacaag ccaaatattt
181 ggaagaacga tttcctagat caatctctta tcagcaaata cgatggagat gagtatcgga
241 agctgtctga gaagttaata gaagaagtta agatttatat atctgctgaa acaatggatt
//
A Traditional
GenBank Record
Header
The Flatfile Format
Feature Table
Sequence
The Header
LOCUS
DEFINITION
ACCESSION
VERSION
KEYWORDS
SOURCE
ORGANISM
REFERENCE
AUTHORS
TITLE
JOURNAL
REFERENCE
AUTHORS
TITLE
JOURNAL
REFERENCE
AUTHORS
TITLE
JOURNAL
REMARK
COMMENT
AY182241
1931 bp
mRNA
linear
PLN 04-MAY-2004
Malus x domestica (E,E)-alpha-farnesene synthase (AFS1) mRNA,
complete cds.
AY182241
AY182241.2 GI:32265057
.
Malus x domestica (cultivated apple)
Malus x domestica
Eukaryota; Viridiplantae; Streptophyta; Embryophyta; Tracheophyta;
Spermatophyta; Magnoliophyta; eudicotyledons; core eudicots;
rosids; eurosids I; Rosales; Rosaceae; Maloideae; Malus.
1 (bases 1 to 1931)
Pechous,S.W. and Whitaker,B.D.
Cloning and functional expression of an (E,E)-alpha-farnesene
synthase cDNA from peel tissue of apple fruit
Planta 219, 84-94 (2004)
2 (bases 1 to 1931)
Pechous,S.W. and Whitaker,B.D.
Direct Submission
Submitted (18-NOV-2002) PSI-Produce Quality and Safety Lab,
USDA-ARS, 10300 Baltimore Ave. Bldg. 002, Rm. 205, Beltsville, MD
20705, USA
3 (bases 1 to 1931)
Pechous,S.W. and Whitaker,B.D.
Direct Submission
Submitted (25-JUN-2003) PSI-Produce Quality and Safety Lab,
USDA-ARS, 10300 Baltimore Ave. Bldg. 002, Rm. 205, Beltsville, MD
20705, USA
Sequence update by submitter
On Jun 26, 2003 this sequence version replaced gi:27804758.
Header: Locus Line
LOCUS
AY182241
1931 bp
mRNA
linear
PLN 04-MAY-2004
DEFINITION
Malus x domestica
synthase (AFS1)
mRNA,
LOCUS
AY182241
1931 (E,E)-alpha-farnesene
bp
mRNA
linear
PLN 04-MAY-2004
complete cds.
ACCESSION
AY182241
VERSION
AY182241.2 GI:32265057
KEYWORDS
.
SOURCE
Malus x domestica (cultivated apple)
ORGANISM Malus x domestica
Eukaryota; Viridiplantae; Streptophyta; Embryophyta; Tracheophyta;
Spermatophyta; Magnoliophyta; eudicotyledons; core eudicots;
rosids; eurosids I; Rosales; Rosaceae; Maloideae; Malus.
REFERENCE
1 (bases 1 to 1931)
AUTHORS
Pechous,S.W. and Whitaker,B.D.
TITLE
Cloning and functional expression of an (E,E)-alpha-farnesene
synthase cDNA from peel tissue of apple fruit
JOURNAL
Planta 219, 84-94 (2004)
REFERENCE
2 (bases 1 to 1931)
AUTHORS
Pechous,S.W. and Whitaker,B.D.
TITLE
Direct Submission
JOURNAL
Submitted (18-NOV-2002) PSI-Produce Quality and Safety Lab,
USDA-ARS, 10300 Baltimore Ave. Bldg. 002, Rm. 205, Beltsville, MD
20705, USA
REFERENCE
3 (bases 1 to 1931)
AUTHORS
Pechous,S.W. and Whitaker,B.D.
TITLE
Direct Submission
JOURNAL
Submitted (25-JUN-2003) PSI-Produce Quality and Safety Lab,
USDA-ARS, 10300 Baltimore Ave. Bldg. 002, Rm. 205, Beltsville, MD
20705, USA
REMARK
Sequence update by submitter
COMMENT
On Jun 26, 2003 this sequence version replaced gi:27804758.
Length
Locus name
Molecule type
Division
Modification Date
Header: Database Identifiers
LOCUS
DEFINITION
AY182241
1931 bp
mRNA
linear
PLN 04-MAY-2004
Malus x domestica (E,E)-alpha-farnesene synthase (AFS1) mRNA,
Accession
complete cds.
ACCESSION
AY182241
•Stable
VERSION
AY182241.2 GI:32265057
•Reportable
KEYWORDS
.
•Universal
SOURCE
Malus x domestica (cultivated apple)
ORGANISM Malus x domestica
Eukaryota; Viridiplantae; Streptophyta; Embryophyta; Tracheophyta;
Spermatophyta; Magnoliophyta; eudicotyledons; core eudicots;
rosids; eurosids I; Rosales; Rosaceae; Maloideae; Malus.
Version
REFERENCE
1 (bases 1 to 1931)
GI number
AUTHORS
Pechous,S.W.
and
Whitaker,B.D.
Tracks changes in sequence
NCBI
internal
use
TITLE
Cloning and functional expression
of an
(E,E)-alpha-farnesene
synthase cDNA from peel tissue of apple fruit
JOURNAL
Planta 219, 84-94 (2004)
REFERENCE
2 (bases 1 to 1931)
AUTHORS
Pechous,S.W. and Whitaker,B.D.
TITLE
Direct Submission
JOURNAL
Submitted (18-NOV-2002) PSI-Produce Quality and Safety Lab,
USDA-ARS, 10300 Baltimore Ave. Bldg. 002, Rm. 205, Beltsville, MD
20705, USA
REFERENCE
3 (bases 1 to 1931)
AUTHORS
Pechous,S.W. and Whitaker,B.D.
TITLE
Direct Submission
JOURNAL
Submitted (25-JUN-2003) PSI-Produce Quality and Safety Lab,
USDA-ARS, 10300 Baltimore Ave. Bldg. 002, Rm. 205, Beltsville, MD
20705, USA
REMARK
Sequence update by submitter
COMMENT
On Jun 26, 2003 this sequence version replaced gi:27804758.
ACCESSION
AY182241
VERSION
AY182241.2
GI:32265057
Header: Organism
LOCUS
DEFINITION
AY182241
1931 bp
mRNA
linear
PLN 04-MAY-2004
Malus x domestica (E,E)-alpha-farnesene synthase (AFS1) mRNA,
complete cds.
ACCESSION
AY182241
VERSION
AY182241.2 GI:32265057
KEYWORDS
.
SOURCE
Malus x domestica (cultivated apple)
SOURCE
(cultivated apple)
ORGANISMMalus
Malusxx domestica
domestica
ORGANISM Malus
x domestica
Eukaryota;
Viridiplantae; Streptophyta; Embryophyta; Tracheophyta;
Spermatophyta;
Magnoliophyta; Streptophyta;
eudicotyledons; core
eudicots;
Eukaryota;
Viridiplantae;
Embryophyta;
rosids; eurosids I; Rosales; Rosaceae; Maloideae; Malus.
Tracheophyta;
Spermatophyta; Magnoliophyta; eudicotyledons;
REFERENCE
1 (bases 1 to 1931)
eudicots;
eurosids I; Rosales; Rosaceae;
AUTHORS core
Pechous,S.W.
androsids;
Whitaker,B.D.
TITLE Maloideae;
Cloning andMalus.
functional expression of an (E,E)-alpha-farnesene
synthase cDNA from peel tissue of apple fruit
JOURNAL
Planta 219, 84-94 (2004)
REFERENCE
2 (bases 1 to 1931)
AUTHORS
Pechous,S.W. and Whitaker,B.D.NCBI-controlled taxonomy
TITLE
Direct Submission
JOURNAL
Submitted (18-NOV-2002) PSI-Produce Quality and Safety Lab,
USDA-ARS, 10300 Baltimore Ave. Bldg. 002, Rm. 205, Beltsville, MD
20705, USA
REFERENCE
3 (bases 1 to 1931)
AUTHORS
Pechous,S.W. and Whitaker,B.D.
TITLE
Direct Submission
JOURNAL
Submitted (25-JUN-2003) PSI-Produce Quality and Safety Lab,
USDA-ARS, 10300 Baltimore Ave. Bldg. 002, Rm. 205, Beltsville, MD
20705, USA
REMARK
Sequence update by submitter
COMMENT
On Jun 26, 2003 this sequence version replaced gi:27804758.
The Feature Table
FEATURES
source
gene
CDS
start (atg)
Coding sequence
Implied
protein
Location/Qualifiers
1..1931
/organism="Malus x domestica"
/mol_type="mRNA"
/cultivar="'Law Rome'"
/db_xref="taxon:3750"
/tissue_type="peel"
1..1931
/gene="AFS1"
stop (tag)
54..1784
/gene="AFS1"
/note="terpene synthase"
/codon_start=1
/product="(E,E)-alpha-farnesene synthase"
/protein_id="AAO22848.2"
GenPept Identifiers
/db_xref="GI:32265058"
/translation="MEFRVHLQADNEQKIFQNQMKPEPEASYLINQRRSANYKPNIWK
NDFLDQSLISKYDGDEYRKLSEKLIEEVKIYISAETMDLVAKLELIDSVRKLGLANLF
EKEIKEALDSIAAIESDNLGTRDDLYGTALHFKILRQHGYKVSQDIFGRFMDEKGTLE
NHHFAHLKGMLELFEASNLGFEGEDILDEAKASLTLALRDSGHICYPDSNLSRDVVHS
LELPSHRRVQWFDVKWQINAYEKDICRVNATLLELAKLNFNVVQAQLQKNLREASRWW
ANLGIADNLKFARDRLVECFACAVGVAFEPEHSSFRICLTKVINLVLIIDDVYDIYGS
EEELKHFTNAVDRWDSRETEQLPECMKMCFQVLYNTTCEIAREIEEENGWNQVLPQLT
KVWADFCKALLVEAEWYNKSHIPTLEEYLRNGCISSSVSVLLVHSFFSITHEGTKEMA
DFLHKNEDLLYNISLIVRLNNDLGTSAAEQERGDSPSSIVCYMREVNASEETARKNIK
GMIDNAWKKVNGKCFTTNQVPFLSSFMNNATNMARVAHSLYKDGDGFGDQEKGPRTHI
The Sequence: 99.99% Accurate
ORIGIN
//
1
61
121
181
ttcttgtatc
tcagagttca
aacctgaagc
ggaagaacga
ccaaacatct
cttgcaagct
ctcttacttg
tttcctagat
cgagcttctt
gataatgagc
attaatcaaa
caatctctta
gtacaccaaa
agaaaatttt
gacggtctgc
tcagcaaata
ttaggtattc
tcaaaaccag
aaattacaag
cgatggagat
actatggaat
atgaaacccg
ccaaatattt
gagtatcgga
1741
1801
1861
1921
ggacccacat
aataaatagc
tgtaacgttg
aaaaaaaaaa
cctgtcttta ctattccaac ctcttgtaaa ctagtactca tatagtttga
agcaaaagtt tgcggttcag ttcgtcatgg ataaattaat ctttacagtt
ttgccaaaga ttatgaataa aaagttgtag tttgtcgttt aaaaaaaaaa
a
GenPept: FASTA format
>gi|32265058|gb|AAO22848.2| (E,E)-alpha-farnesene synthase [Malus x domestica]
MEFRVHLQADNEQKIFQNQMKPEPEASYLINQRRSANYKPNIWKNDFLDQSLISKYDGDEYRKLSEKLIE
EVKIYISAETMDLVAKLELIDSVRKLGLANLFEKEIKEALDSIAAIESDNLGTRDDLYGTALHFKILRQH
GYKVSQDIFGRFMDEKGTLENHHFAHLKGMLELFEASNLGFEGEDILDEAKASLTLALRDSGHICYPDSN
LSRDVVHSLELPSHRRVQWFDVKWQINAYEKDICRVNATLLELAKLNFNVVQAQLQKNLREASRWWANLG
IADNLKFARDRLVECFACAVGVAFEPEHSSFRICLTKVINLVLIIDDVYDIYGSEEELKHFTNAVDRWDS
RETEQLPECMKMCFQVLYNTTCEIAREIEEENGWNQVLPQLTKVWADFCKALLVEAEWYNKSHIPTLEEY
LRNGCISSSVSVLLVHSFFSITHEGTKEMADFLHKNEDLLYNISLIVRLNNDLGTSAAEQERGDSPSSIV
CYMREVNASEETARKNIKGMIDNAWKKVNGKCFTTNQVPFLSSFMNNATNMARVAHSLYKDGDGFGDQEK
GPRTHILSLLFQPLVN
>gi|32265070|gb|AAP75563.1| putative doublecortin domain-containing protein
MAKTGAEDHREALSQSSLSLLTEAMEVLQQSSPEGTLDGNTVNPIYKYILNDLPREFMSSQAKAVIKTTD
DYLQSQFGPNRLVHSAAVSEGSGLQDCSTHQTASDHSHDEISDLDSYKSNSKNNSCSISASKRNRPVSAP
VGQLRVAEFSSLKFQSARNWQKLSQRHKLQPRVIKVTAYKNGSRTVFARVTAPTITLLLEECTEKLNLNM
AARRVFLADGKEALEPEDIPHEADVYVSTGEPFLNPFKKIKDHLLLIKKVTWTMNGLMLPTDIKRRKTKP
VLSIRMKKLTERTSVRILFFKNGMGQDGHEITVGKETMKKVLDTCTIRMNLNLPARYFYDLYGRKIEDIS
KGKH
Abstract Syntax Notation: ASN.1
Seq-entry ::= set {
class nuc-prot ,
descr {
title "Malus x domestica (E,E)-alpha-farnesene synthase
(AFS1) mRNA,
complete cds." ,
source {
org {
taxname "Malus x domestica" ,
common "cultivated apple" ,
db {
{
db "taxon" ,
tag
id 3750 } } ,
orgname {
name
binomial {
genus "Malus" ,
species "x domestica" } ,
mod {
{
subtype cultivar ,
GenPept
GenBank
ASN.1
FASTA
Protein
FASTA
Nucleotide
Database 2: RefSeq
NCBI’s Derivative Sequence Database
What is RefSeq?
•
Curated transcripts and proteins (NM_, NP_)
– reviewed
– human, mouse, rat, fruit fly, zebrafish, arabidopsis
•
•
microbial genomes (proteins), and more
Model transcripts and proteins (XM_, XP_)
Assembled Genomic Regions (contigs) (NT_, NW_)
– human genome
– mouse genome
•
– rat genome
Chromosome records (NC_)
– Human genome
– microbial
srcdb_refseq[Properties]
– organelle
ftp://ftp.ncbi.nih.gov/refseq/release/
RefSeq Benefits
•
•
•
•
•
•
•
non-redundancy
explicitly linked nucleotide and protein sequences
updates to reflect current sequence data and biology
data validation
format consistency
distinct accession series
stewardship by NCBI staff and collaborators
RefSeq Curation Processes
Curated genomic DNA
(NC, NT, NW)
Scanning....
Curated Model mRNA (XM)
Model protein (XP)
(XR)
Curated mRNA (NM)
(NR)
Protein (NP)
RefSeq Accession Numbers
mRNAs and Proteins
NM_123456
NP_123456
NR_123456
XM_123456
XP_123456
XR_123456
Gene Records
NG_123456
Chromosome
NC_123455
Assemblies
NT_123456
NW_123456
Curated mRNA
Curated Protein
Curated non-coding RNA
Predicted mRNA
Predicted Protein
Predicted non-coding RNA
Reference Genomic Sequence
Microbial replicons, organelle , viral
genomes, human chromosomes
Contig
WGS Supercontig
From GenBank to RefSeq
NM_000121: Sequence Revision History
Database 3: UniGene
NCBI’s Derivative EST Database
UniGene
Clustering Expressed Sequences
•
•
•
Records are clusters of mRNAs and ESTs that ideally
represent single genes
Records are created automatically by a modified BLAST
algorithm
UniGene provides a means to identify an EST or
unannotated mRNA
UniGene
Gene-oriented clusters of expressed sequences
• Automatic clustering using MegaBlast
• Each cluster represents a unique gene
• Informed by genome hits
• Information on tissue types and map locations
• Useful for gene discovery and selection of mapping
reagents
A Cluster of ESTs
query
5’ EST hits
3’ EST hits
UniGene Collections
Example UniGene Cluster
Histogram of cluster sizes for UniGene Hs Build 177
(Now at Build #186)
UniGene Cluster Hs.95351
SELECTED PROTEIN SIMILARITES
UniGene Cluster Hs.95351
GENE EXPRESSION
UniGene Cluster Hs.95351: expression
UniGene Cluster Hs.95351: seqs
Download sequences
web page
ftp://ftp.ncbi.nih.gov/repository/UniGene/Homo_sapiens/
Database 4: MMDB
NCBI’s derivative protein structure database
Indexing into MMDB
MMDB
Molecular Modeling
Data Base
Structure
• Import only experimentally determined structures
• Convert to ASN.1
• Create “backbone” model (Cα, P only)
• Verify sequences
• Create single-conformer model
Add secondary structure
Add chemical bonds
id 1 ,
name "helix 1" ,
type helix ,
location
subgraph
residues
interval {
{ molecule-id 1 ,
from 49 ,
to 61 } } } ,
inter-residue-bonds {
{
atom-id-1 {
molecule-id 1 ,
residue-id 1 ,
atom-id 1 } ,
atom-id-2 {
molecule-id 1 ,
residue-id 2 ,
atom-id 9 } } ,
Structure Summary
Cn3D viewer
Structure Neighbors
Conserved Domains
3D Domain Neighbors
Cn3D 4.1: C-Src
Cn3D 4.1: Structural
Alignment
Conserved ATP binding site
Src Kinase H. sapiens
Casein kinase S. pombe
Cn3D: Simple Homology Modeling
human
swordtail
NCBI CD: Tyrosine Kinase
Using Cn3D to model domains
Submitting a PDB File to VAST
• Choose the file format
• Remove all lines except
ATOM
This is the best way to convert
PDB files to MMDB format
for viewing with Cn3D!
Database 5: GEO
NCBI’s Gene Expression Omnibus
Submitted by
Manufacturer*
GPL
Platform
descriptions
Submitted by
Experimentalists
GSM
GSE
Grouping of
Raw/processed
slide/chip data
spot intensities
from a single “a single experiment”
slide/chip
GEO SaMple:
GEO SEries:
experimental
set of related
conditions
samples
Entrez GEO
Curated by
NCBI
GDS
Grouping of
experiments
Entrez
GEO Datasets
What’s a DataSet?
Supplied by
submitter
Platform
Sample
Series
(GPL)
(GSM)
(GSE)
array definition
hyb. measurements
related Samples
DataSet
Assembled
by GEO staff
(GDS)
• A collection of experimentally-related samples
processed using the same platform.
• Samples within DataSets are organized into
subgroups based on experimental variables.
• Form the basis of GEO’s query, analysis and data
display tools.
Gene Expression Omnibus
Dataset browser
GEO Dataset Browser
GEO Dataset Report
GEO Profiles
… of 12625
Database 6: CDD
NCBI’s Derivative Conserved Domain
Database
Entrez CDD
•
•
•
Conserved Domain
Database
Multiple sequence alignments
Position-specific scoring matrices (PSSM)
Sources SMART, PFAM, COGs, KOGs, and
NCBI curated domains (structure-informed alignments)
CDD
>gi|45549418|gb|AAS67634.1| ATP7A [Solenodon paradoxus]
IVYQPHLITVEEIKKQIKAVGFPAFIKKQPKYLKLGAIDIERLKNIPVKSSEGSQQMSPS
STNDSKVTLTIDGMHCNSCVSNIESALSTLHYVSSIVVSLQNKSAIIKYNANSVTPEIL
KKAIEAISPGQYRVSITSEVESTSNSPSSSSQKAPLNVVSQPLTQVTVININGMTCNS
CVQSIEGVMSKKAGVKSIQVSLANRNGTVEYDP LLTSPEILRE
CDD
Click on a colored bar to align your sequence to
the CD
CD
Pfam
COG
Conserved Domain Database: cd00371.1, HMA
CDD
CDART: Conserved Domain Architecture
Retrieval Tool
Database 7: NCBI Genome Map
Viewing Complex Genomes
NCBI Map Viewer
• Map Viewer Home Page
• Shows all supported organisms
• Provides links to genomic BLAST
– Genome Overview Page
• Provides links to individual chromosomes
• Shows hits on a genome graphically
– Chromosome Viewing Page
• Allows interactive views of annotation details
• Provides numerous maps unique to each genome
The Map Viewer
Genome BLAST
Map Viewer: Human MLH1
EST Hits
Customizable
Transcripts
Models
NCBI Assembly
Gene Annotations
Maps and Options
Mapped Variations
MLH1 Synteny: Mammalian
Genomes
Many Other NCBI Databases…
Other Specialized Databases
•
•
•
•
•
•
Gene Symbol Database ( HUGO Gene Nomenclature )
KEGG (Kyoto Encyclopedia of Genes and Genomes)
Pathway
EPD (Eukaryote Promoter Database)
Transcription Factor Database ( TRANSFAC )
Many organism-specific databases (e.g., Flybase,
Beebase)
…
Access Databases through
Entrez
Accessing the Data in Entrez
•
Web Tools
– Batch Entrez
• Upload a file of GI or accession numbers to retrieve sequences
– Batch Citation Matcher
• Send citation information to Entrez and retrieve PubMed IDs for
linking, citation display or other applications
– Advanced Entrez Searching
• Advanced searching techniques for Web Entrez
– My NCBI
• Includes automatic e-mailing of search updates and filters for search
results
•
• Requires a username and password to access stored searches
Programming Tools
– E-Utilities
• Run Entrez queries and download data from your own scripts over the
Web
– Linking to Entrez
• Link to specific Entrez pages from your own web pages or applications
– Entrez Client/Server
• C language library for embedding Entrez calls into your programs
Entrez: Web Access
Default search: Against all databases in Entrez
Interface: Global Entrez
Target database: Adjustable using the pull-down menu
NCBI Toolbox
/************************************************************************
*
*
asn2ff.c
*
convert an ASN.1 entry to flat file format, using the FFPrintArray.
*
**************************************************************************/
#include <accentr.h>
#include "asn2ff.h"
#include "asn2ffp.h"
#include "ffprint.h"
#include <subutil.h>
#include <objall.h>
#include <objcode.h>
#include <lsqfetch.h>
#include <explore.h>
Toolbox Sources
ftp> open ftp.ncbi.nih.gov
.
.
#ifdef ENABLE_ID1
ftp> cd toolbox
#include <accid1.h>
#endif
ftp> cd ncbi_tools
FILE *fpl;
ftp://ftp.ncbi.nlm.gov/toolbox/ncbi_tools
Args myargs[] = {
{"Filename for asn.1 input","stdin",NULL,NULL,TRUE,'a',ARG_FILE_IN,0.0,0,NULL},
{"Input is a Seq-entry","F", NULL ,NULL ,TRUE,'e',ARG_BOOLEAN,0.0,0,NULL},
{"Input asnfile in binary mode","F",NULL,NULL,TRUE,'b',ARG_BOOLEAN,0.0,0,NULL},
{"Output Filename","stdout", NULL,NULL,TRUE,'o',ARG_FILE_OUT,0.0,0,NULL},
{"Show Sequence?","T", NULL ,NULL ,TRUE,'h',ARG_BOOLEAN,0.0,0,NULL},
Challenges in Bioinformatics
PMC
Cancer Chromosome
OMIM
Word weight
Books
PubMed
Abstracts
Related Articles
PubChem
How can we help biologists
Taxonomy
manage and exploit all suchVAST
3 -D
Structure
rapid growing, heterogeneous,
and
Gene
Genome
inaccurate information both
efficiently and effectively?
3D domain
CDD
Phylogeny
Neighbors
Related Structures
HomoloGene
Genome Project
UniGene
GEO
SNP
OMIM
BLAST
BLAST
Protein
Sequences
Nucleotide
Sequences
Neighbors
Related Sequences
Hard Link
Neighbors
Related Sequences
BLink
Domains