Document
advertisement

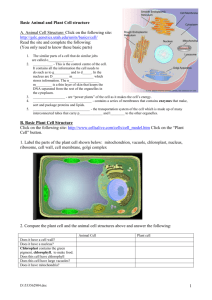
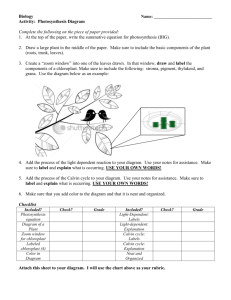
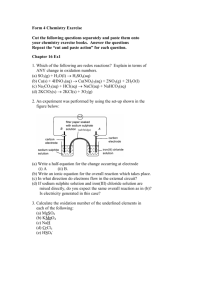
Genetics 2120 Sequence Analysis Extra Credit Assignment – Spring 2005 Genetics 2120 Genome Project – Part VI Interpreting sequence information Our goal is to eventually sequence the entire Fescue chloroplast genome. What we need initially is a set of clues which will enable us to take these first sequences and fragments and begin the process of piecing it all together. To do this we have to make some assumptions about our experimental genome. All of the initial design was based on the maize chloroplast genome therefore I will ask you to make all initial comparisons based on that one. Some of your searches will tell you that other plants may be more related to Fescue than maize, which is important in the long haul, but for the moment we need to get an idea of how to build our map and it’s easiest to stick with a single reference. Please keep in mind that we have no idea what we’ll find and there is no guarantee that any clone will even be a piece of the chloroplast genome. Random pieces could have been amplified and cloned. Also, portions of the genome will be arranged in ways no one can predict. If you would like an analogy… We are attempting to piece together a clear landscape photograph while using an impressionistic painting of a similar landscape as your reference picture (happy clouds included). STEP 1: Is the sequence homologous to any other known sequences? Welcome to the world of Bioinformatics. (note the lack of an exclamation point) Go to the National Center for Biotechnology Information website. http://www.ncbi.nlm.nih.gov/ We want to perform a BLAST (Basic Local Alignment Search Tool) search. BLAST is a program which compares DNA and protein sequences and aligns them by similarity. Two similar sequences are called homologues. Discerning geneticists choose BLAST 1 Genetics 2120 Sequence Analysis Extra Credit Assignment – Spring 2005 You want to compare your DNA sequence to others so choose a nucleotide-nucleotide BLAST (blastn). Choose Nucleotide-nucleotide BLAST (blastn) You were provided a sequence in a text file. Open the file using Word; highlight the sequence, with your cursor in the highlighted area press the right-hand button (right-click) on the mouse so a menu pops up, click copy from the list of commands. Back at the NCBI page left-click inside the Search box and then right-click to open the list of commands, choose Paste. For the initial search I want you to only search against the corn database (Zea mays). 1. Paste your sequence here 3. Click on BLAST! button 2. Choose Zea mays [org] from the pull-down menu. 2 Genetics 2120 Sequence Analysis Extra Credit Assignment – Spring 2005 You will get a formatting screen. We will use the default settings. Before you click on the Format! button, I want you to copy the request ID number and paste it into the report you will hand in. I want specific information in your report so follow the format I outline below. How to Make Your Report Document 1. 2. 3. 4. 5. Start a new document in Word. At the top of the first page put your name and the name of the sequence you are analyzing. Paste the sequence you are analyzing in your document. Skip a few lines and make a subject heading called “Zea mays Sequence Homology Results” Copy and paste the request ID number from your formatting page. Press Format! and wait When the search is completed the results page will pop up in place of the formatting page. Scroll down until you get an image similar to the one shown below. Inside the boxed area is a number line, this represents your piece of DNA. The colored lines below represent pieces of DNA that are similar to your piece. Below the boxed area is a list of the homologues. The most likely hit will be the complete chloroplast genome of Zea mays. If this is not listed you either have something very interesting or a mistake was made in the cloning process. 3 Genetics 2120 Sequence Analysis Extra Credit Assignment – Spring 2005 If you have a chloroplast homologue scroll down until you get to the actual base for base lineups like the one shown below (if you don’t have a chloroplast homologue then skip to the portion below where it says, “Search the Whole Database”). Find the lineup that has this header… >gi|11990232|emb|X86563.2|ZMA86563 Zea mays complete chloroplast genome Length = 140384 The two sequences in the lineup are the one you entered (Query) and its closest match (Sbjct). Your Report Document 6. Copy the text starting with the header and ending with the final base pairing and paste it in your report document below the request ID. Search the Whole Database Now repeat your search using the whole database. This time when you enter your sequence do not limit your search to Zea mays, instead allow the program to search all organisms (this is the default setting so you just enter your sequence and press BLAST!). Your Report Document 7. Make another subject heading called “Whole Database Homology Search”. 8. Copy the request ID. The results page may resemble the one shown on the right. You may notice that the top hit is not Zea mays but Oryza sativa (rice). 4 Genetics 2120 Sequence Analysis Extra Credit Assignment – Spring 2005 Your Report Document 9. Just under the box with the number line the similar sequences are listed. Copy the top 5 (or so) and paste them into your report document. 10. If you didn’t get a maize chloroplast homologue… but you did get a chloroplast homologue from another species then go to the base-for base lineup described above (Step 6 for “Your Report Document”) then copy and paste that sequence into your report. What if my sequence doesn’t have a chloroplast homologue? If there are no chloroplast homologues at all take a close look at the matches that do appear. The BLAST program will try to make your sequence fit other sequences even if they have very little in common. For this reason another number (the E-value) is included so that you (the human with the critical thinking skills) can determine if the match represents a bona fide homologous region or was forced by the program. The E-value that is assigned to each potential homologue represents number of times your database match may have occurred by random chance. The lower the e-value the more confident you can be that you are looking at two similar sequences with a common ancestry. Typically, Evalues equal or greater than 10-4 are not similar enough to represent true homologues. If you get a match with human DNA that is less than 10-4 then we probably cloned a contaminant (a piece of DNA from me or someone else from the class). No matter what you find, copy and paste the top hits into your report so the analysis can continue. Who knows, it may actually be a part of the fescue genome! Your Report Document 11. What is the significance of any similar regions you found either chloroplast or nonchloroplast? 5 Genetics 2120 Sequence Analysis Extra Credit Assignment – Spring 2005 STEP 2: Are there any genes in your sequence? The first step is interesting and essential in our analysis but the genes are the most exciting part. A variant of BLAST called BlastX will take your sequence, automatically translate it using every possible reading frame in both directions, and then compare these hypothetical proteins to every known protein. From the BLAST page choose the Translated query vs. protein database (blastx). Choose Translated query vs. protein database (blastx) Perform the same steps you did with the blastn search before. Note: If you found maize chloroplast hits before then you want to begin by limiting your search to the maize database using the pull-down menu under “Options”. After that search, repeat the search using all species. If you found hits to another species you should limit your search to that species’ database before searching the entire database. Key to monocot species names. Oryza sp. – Rice Triticum sp. – Wheat Saccharum sp. – Sugarcane Zea mays - Corn Your Report Document 12. Start a new subject heading titled “BlastX homology search”. 13. For each search include the Request ID, and the top matches (use the same criterion of an E-value less than 10-4). 6 Genetics 2120 Sequence Analysis Extra Credit Assignment – Spring 2005 Your results page will resemble this one. Notice that the lineup is now between amino acids instead of nucleotides. There may be pieces of more than one gene on the fragment you are analyzing. Be sure to include all of the hits in your report. Your Report Document 14. What is the significance of any homologues you found during the searches? 7 Genetics 2120 Sequence Analysis Extra Credit Assignment – Spring 2005 Report Document Checklist Summary of Report Document Requests, Include… General Info. 1. Start a new document in Word. 2. At the top of the first page put your name and the name of the sequence you are analyzing. 3. Paste the sequence you are analyzing in your document. BLAST Search Results 4. Skip a few lines and make a subject heading called “Zea mays Sequence Homology Results” 5. Copy and paste the request ID number from your formatting page. 6. Copy the text starting with the header and ending with the final base pairing and paste it in your report document below the request ID. 7. Make another subject heading called “Whole Database Homology Search”. 8. Copy the request ID. 9. Just under the box with the number line the similar sequences are listed. Copy the top 5 (or so) and paste them into your report document. 10. If you didn’t get a maize chloroplast homologue… but you did get a chloroplast homologue from another species then go to the base-for base lineup described above (Step 6 for “Your Report Document”) then copy and paste that sequence into your report. 11. What is the significance of any similar regions you found either chloroplast or nonchloroplast? BLASTX Search Results 12. Start a new subject heading titled “BlastX homology search”. 13. For each search include the Request ID, and the top matches (use the same criterion of an E-value less than 10-4). 14. What is the significance of any homologues you found during the searches? Your report is DUE APRIL 15TH BY 3:00 P.M. Late submissions will be accepted but no credit will be awarded. 8