Jenae`s BLAST search 10-23-12
advertisement
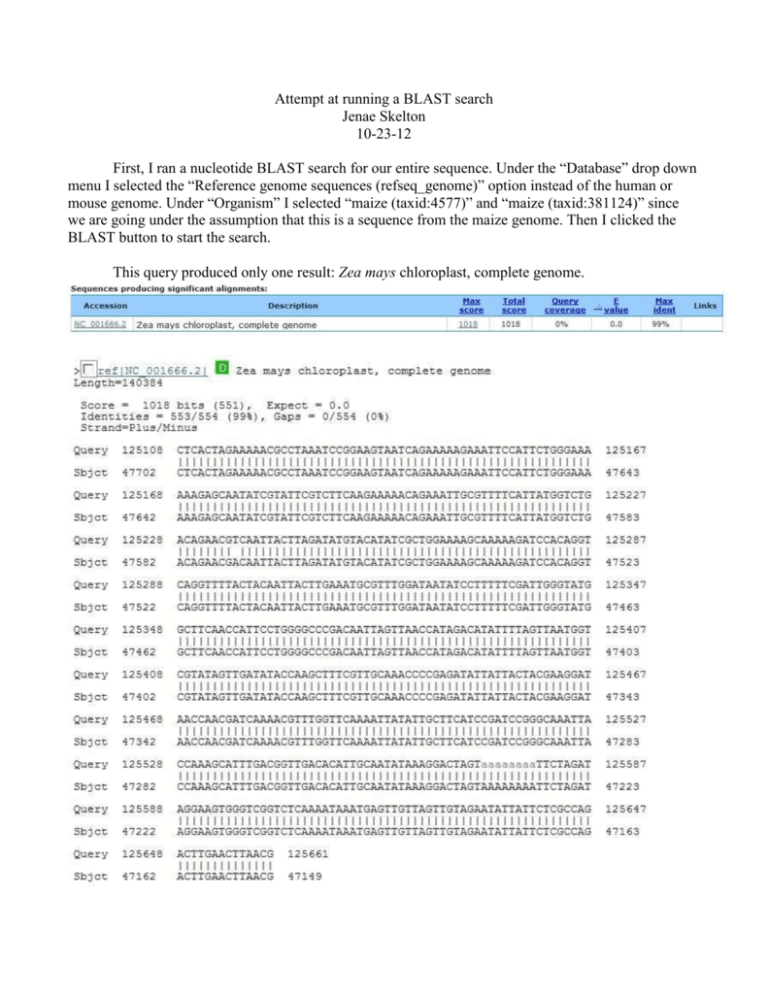
Attempt at running a BLAST search Jenae Skelton 10-23-12 First, I ran a nucleotide BLAST search for our entire sequence. Under the “Database” drop down menu I selected the “Reference genome sequences (refseq_genome)” option instead of the human or mouse genome. Under “Organism” I selected “maize (taxid:4577)” and “maize (taxid:381124)” since we are going under the assumption that this is a sequence from the maize genome. Then I clicked the BLAST button to start the search. This query produced only one result: Zea mays chloroplast, complete genome. Then I ran a protein BLAST search for our entire sequence. I used the “Non-redundant protein sequences (nr)” database and the same organism categories as the previous search. This search failed to run and only gave an error message that stated “Message ID#24 Error: Failed to read the Blast query: Nucleotide FASTA provided for protein sequence.” I realized after the fact that I should have done a BLASTx search since I was looking for proteins using a nucleotide sequence. So I went back to the original BLASTn results and opened the accession number in a new window. I clicked on the “Run BLAST” on the side bar under “Analyze This Sequence.” This produced a list of 170 Blast hits. This is what I have so far. I have no idea if I even did any of this correctly. I have no idea what the next step should be. I don’t know if this is helpful to any of you, but I wanted to play around with the BLAST search to see if I could even run it. Since this is possibly the chloroplast genome, do we need to list all the proteins encoded in it? I don’t know where we should go from here, or if this is even the right analysis and results we should have received.