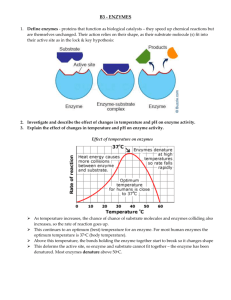
Restriction_Maps
advertisement
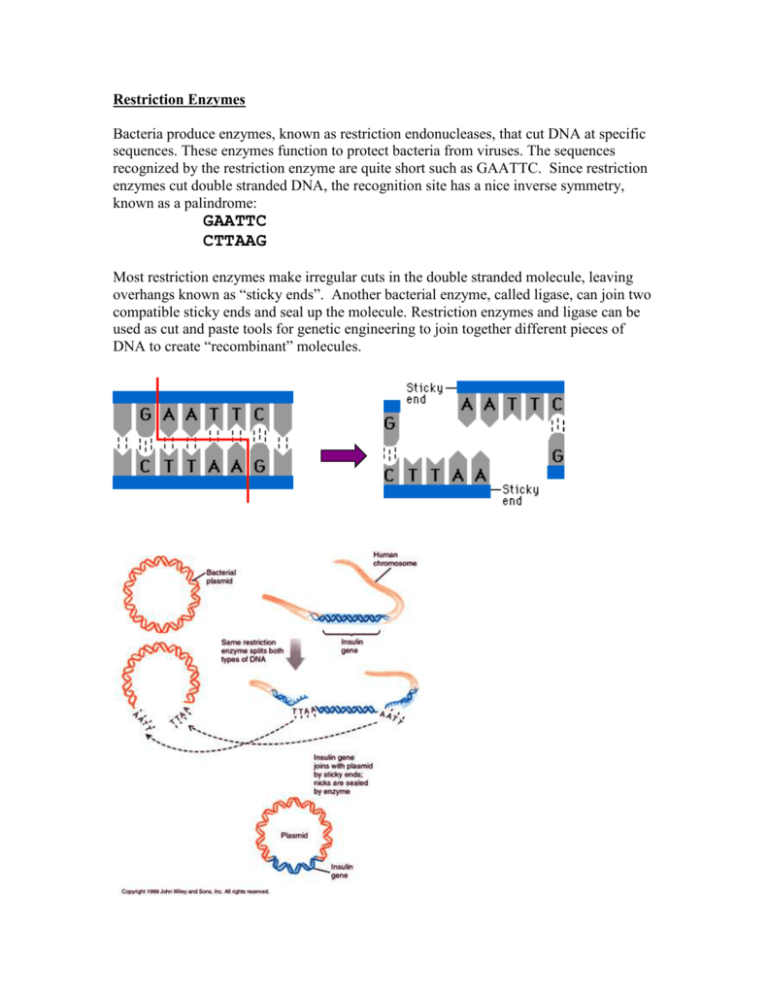
Restriction Enzymes Bacteria produce enzymes, known as restriction endonucleases, that cut DNA at specific sequences. These enzymes function to protect bacteria from viruses. The sequences recognized by the restriction enzyme are quite short such as GAATTC. Since restriction enzymes cut double stranded DNA, the recognition site has a nice inverse symmetry, known as a palindrome: GAATTC CTTAAG Most restriction enzymes make irregular cuts in the double stranded molecule, leaving overhangs known as “sticky ends”. Another bacterial enzyme, called ligase, can join two compatible sticky ends and seal up the molecule. Restriction enzymes and ligase can be used as cut and paste tools for genetic engineering to join together different pieces of DNA to create “recombinant” molecules. http://fig.cox.miami.edu/~cmallery/150/gene/splicing.jpg Over 3500 restriction enzymes have been discovered (about 600 of them are commercially available from various manufacturers), with a total of 253 distinct recognition sites. It is an interesting bioinformatics problem to scan a DNA sequence for restriction recognition sites. Often there are constraints, such as the need to find the same site on two different pieces of DNA that will by joined (or sites with compatible sticky ends), or to find sites that can be cut by a set of enzymes that are available in the lab. Many software tools have been written to solve this simple pattern search problem. As a demonstration, you can use the “Find” function of your web browser to do a simple search for BamHI sites in the human insulin gene. Get the sequence of human insulin from the UCSC genome browser (INS_HUMAN on chromosome 11; see Genome Browser tutorial). Get the genomic sequence (including introns, CDS Exons, 5’ and 3’ UTR Exons) on your screen. Now choose “Find” from the “Edit” menu of your browser and type in GGATCC (the recognition site for BamH1 enzyme). If you had just 3 or 4 enzymes in your freezer, then this process would be simple enough. However, if you needed to find an enzyme site in a very small piece of DNA, then you might want to screen all possible enzymes, and that would take a long time to search for the sequences one by one. New England Biolabs has a nice web tool called NEBcutter (http://tools.neb.com/NEBcutter2) that screens a DNA sequence against all enzymes. Webcutter is another similar program: http://www.firstmarket.com/cutter/cut2.html. NEBcutter2 has some nice features. Once you have pasted in a DNA sequence (use the INS_HUMAN sequence again) and hit the submit button, it produces a graphical restriction map, showing where each enzyme cuts. By default, the display shows only enzymes that cut the sequence a single time, but it can be adjusted with the Display menu to show enzymes that cut 2 or 3 times (2 cutters and 3 cutters). QuickTi me™ and a TIFF (LZW) decompressor are needed to see this picture. Each enzyme shown on the map is linked to a page of data describing its recognition site and chemical properties, the location of its cut site(s) in your sequence, and a list of other enzymes that produce compatible ends that might be used to ligate a fragment cut with this enzyme. QuickTime™ and a TIFF (LZW) decompressor are needed to see this picture. NEBcutter2 will allow you to pick a specific set of enzymes (with the Custom digest option) to cut your sequence and show the resulting fragments in a map, a table of fragment sizes, and a simulated agarose gel electrophoresis image. QuickTime™ and a TIFF (LZW) decompressor are needed to see this picture. There are many uses for such a restriction enzyme analysis tool. In order to clone a fragment of DNA, a scientist must first determine which restriction enzyme (or combination of enzymes) to use. The chart of compatible ends may be useful if the fragment is to be cloned into a vector such as a plasmid that has a multiple cloning site that contains sites for a limited number of enzymes. The table of predicted fragment sizes and the simulated gel image are useful to check if a combination of enzymes will produce several fragments of convenient sizes, and if it is possible to adjust the agarose concentration of a gel in order to separate them.