Lab # 7 Restriction Enzymes
advertisement
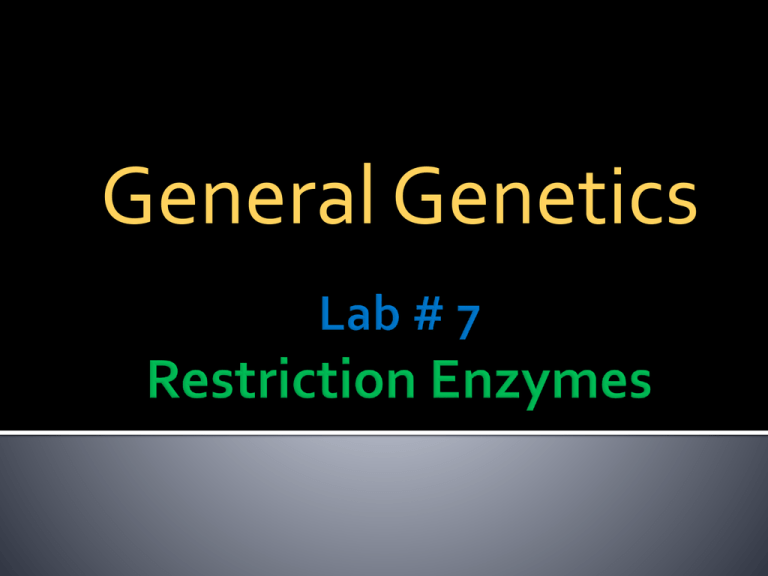
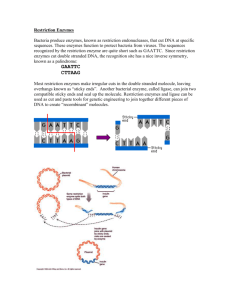
General Genetics 1. Introduce the students to digest genomic DNA by restriction endonucleases. 2. Observe the results of digestion on agarose gel electrophoresis. 1. 2. 3. 4. The activity of restriction enzymes is dependent upon precise environmental conditions: PH Temperature Salt Concentration Ions An Enzymatic Unit (u) is defined as the amount of enzyme required to digest 1 ug of DNA under optimal conditions: 3-5 u/ug of genomic DNA 1 u/ug of plasmid DNA Stocks typically at 10 u/ul Cohesive Ends Cohesive Ends Blunt Ends (5´ Overhang) (3´ Overhang) (No Overhang) BamH1 KpnI HaeIII GGATCC CCTAGG GGTACC CCATGG GGCC CCGG Hundreds of restriction enzymes have been identified. Most recognize and cut palindromic sequences Many leave staggered (sticky) ends by choosing correct enzymes can cut DNA very precisely Important for molecular biologists because restriction enzymes create unpaired "sticky ends" which anneal with any complementary sequence Bacterial" immune system": destroy any "non-self" DNA methylase recognizes same sequence in host DNA and protects it by methylating it; restriction enzyme destroys unprotected = non-self DNA (restriction/modification systems) As an example, consider a 5000 base pair, circular plasmid DNA containing single recognition sites for enzymes A, B, and C. Any one of these enzymes will cleave the DNA once to produce a linear molecule of 5000 base pairs. Differently paired combinations of enzymes in the same reaction mixture (double-digests) will produce the following DNA fragments (sizes in base pairs): •Arbitrarily placing one of the cleavage sites at the top of a circle. This site acts as a reference point. •The closest cleavage site to this point can be placed in a clockwise or counterclockwise direction. Generally, a restriction enzyme map is constructed by first determining the number of fragments each individual enzyme produces. The size and number of fragments is determined by electrophoresis. If a DNA molecule contains several recognition sites for a restriction enzyme, then under certain experimental conditions, it is possible that certain sites are cleaved but not others. These incompletely cleaved fragments of DNA are called partial digests (partials). Partials can arise if an insufficient amount of enzyme is used or the reaction is stopped after a short time (Figure 5). Reactions containing partials may also contain some molecules that have been completely cleaved. Two possible maps inferred from the observations BamH1 XhoI 4.3 kb 3.7 kb 2.3 kb 1.9 kb 1.4 kb 1.3 kb 0.7 kb BamH1 XhoI PCR and Restriction enzymes