SECTION I. User Information
advertisement
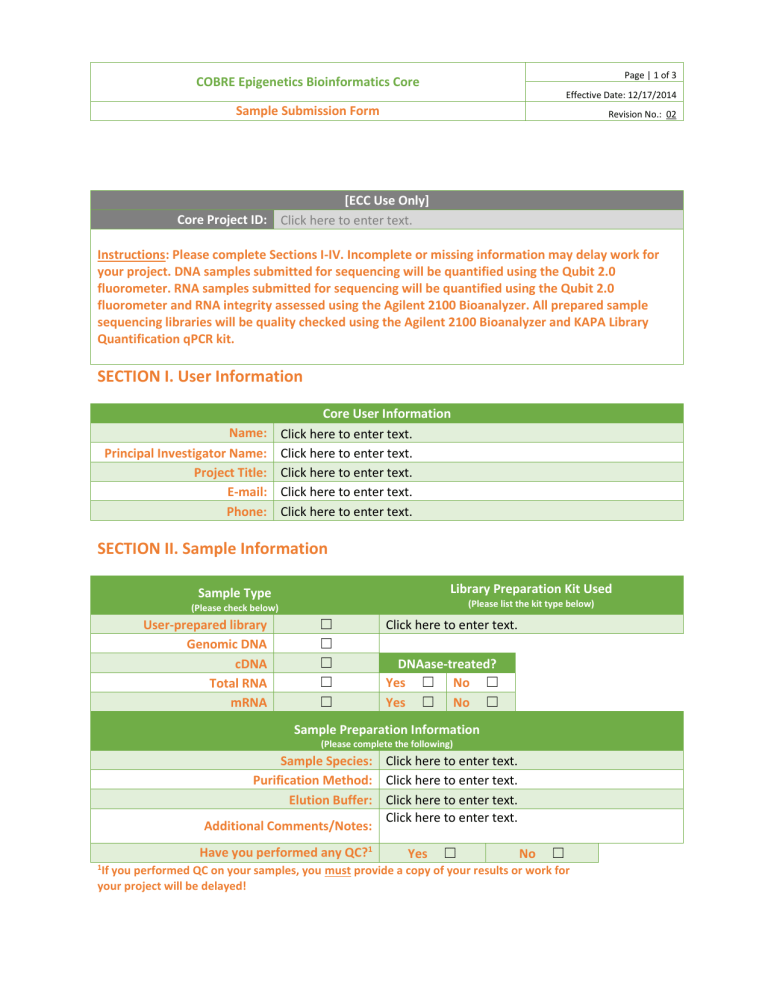
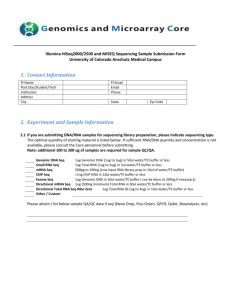
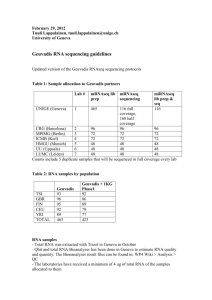
Page | 1 of 3 COBRE Epigenetics Bioinformatics Core Effective Date: 12/17/2014 Sample Submission Form Revision No.: 02 [ECC Use Only] Core Project ID: Click here to enter text. Instructions: Please complete Sections I-IV. Incomplete or missing information may delay work for your project. DNA samples submitted for sequencing will be quantified using the Qubit 2.0 fluorometer. RNA samples submitted for sequencing will be quantified using the Qubit 2.0 fluorometer and RNA integrity assessed using the Agilent 2100 Bioanalyzer. All prepared sample sequencing libraries will be quality checked using the Agilent 2100 Bioanalyzer and KAPA Library Quantification qPCR kit. SECTION I. User Information Name: Principal Investigator Name: Project Title: E-mail: Phone: Core User Information Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. SECTION II. Sample Information Library Preparation Kit Used Sample Type (Please list the kit type below) (Please check below) User-prepared library Genomic DNA cDNA Total RNA mRNA ☐ ☐ ☐ ☐ ☐ Click here to enter text. DNAase-treated? Yes ☐ No ☐ Yes ☐ No ☐ Sample Preparation Information (Please complete the following) Sample Species: Click here to enter text. Purification Method: Click here to enter text. Elution Buffer: Click here to enter text. Click here to enter text. Additional Comments/Notes: Have you performed any QC?1 1 Yes ☐ No ☐ If you performed QC on your samples, you must provide a copy of your results or work for your project will be delayed! Page | 2 of 3 COBRE Epigenetics Bioinformatics Core Effective Date: 12/17/2014 Sample Submission Form Revision No.: 02 SECTION III. Application Application Type (Please check below) RNA-Seq2 ChIP-Seq Small RNA Sequencing Small Genome Sequencing Infinium Methylation Array3 ☐ ☐ ☐ ☐ ☐ 2 Only small genomes (i.e. Bacteria/Fungi) can be sequenced on the MiSeq. Larger genomes (i.e. C.Elegans/Drosophila/Human/Mouse) will be sent to UMGC for sequencing on the HiSeq. 3 Infinium Methylation Arrays will be sent to UMGC. Library Prep: TruSeq RNA TruSeq Small RNA TruSeq ChIP TruSeq DNA PCR-Free Nextera XT DNA Nextera DNA Other (List below) ☐ ☐ ☐ ☐ ☐ ☐ ☐ [ECC Use Only] Sequencing Run Type Single Read ☐ Paired End Read ☐ Sequencing Run Length: 75 bp ☐ 150 bp ☐ 250 bp ☐ 300 bp ☐ Reagent Kit: V2 50 cycles V2 300 cycles V2 500 cycles Micro V2 300 cycles Nano V2 300 cycles Nano V2 500 cycles V3 150 cycles V3 600 cycles ☐ ☐ ☐ ☐ ☐ ☐ ☐ ☐ Page | 3 of 3 COBRE Epigenetics Bioinformatics Core Effective Date: 12/17/2014 Sample Submission Form Revision No.: 02 SECTION IV. Sample List NOTE: For DNA samples, please submit ≥ 1 µg of high quality DNA, concentration > 50 ng/µL; for RNA samples, please submit ≥ 1 µg of total RNA, concentration of at least 50 ng/µL [ECC Use Only] Sample Name Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Click here to enter text. Concentration (ng/µL) Sample Volume (µL) Adapter Index Used (User-prepared libraries only)