NPH_4164_sm_MethodsS1
advertisement

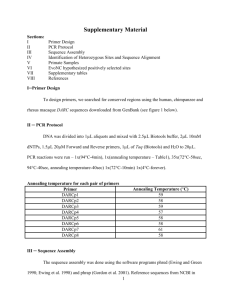
Methods S1 Sequence trimming Newly generated chromatogram files were exported to PHRED for base calling (Ewing & Green, 1998; Ewing et al., 1998). Only inserts longer than 100 bp, with a Phred score >20 that did not belong to a low complexity area were exported into a FASTA format with the corresponding quality file. Publicly available M. incognita ESTs were retrieved from NCBI dbEST. Inserts were detected and cleaned from contaminating sequences (vector, adaptator, poly(A) or poly(T) tails and repeat) (http://www.phrap.org/phredphrap/general.html) using and cross_match RepeatMasker Version 0.990329 Version 3.1.9. (http://repeatmasker.org). Additional extremity trimming was made using the UNIX command-line "trimseq" from European Molecular Biology Open Software Suite (EMBOSS) (Rice et al., 2000). EST datasets were cleared from plant contaminants by blastn (Altschul et al., 1990) against the GenBank nr database and elimination of sequences identical to plant sequences with Expect value=0. In situ hybridizations In situ hybridizations were performed as described by (de Boer et al., 1998) with the following modifications: parasitic juveniles were fixed with 4% formaldehyde (SigmaAldrich, Steinheim, Germany) in Phosphate Buffer Saline (PBS) for 2 days at room temperature. Nematodes were cut into sections and permeabilized with 1 mg/ml proteinase K (Applied Biosystems/Ambion, Austin, TX, USA) for 1 hour at 37°C. After three washes in PBS, sections of nematodes were pelleted, frozen at -80°C for 15 min, incubated at room temperature in cold methanol for 2 min and in acetone for 15 min at -80°C. After centrifugation at 10 000 rpm for 1 min, nematode sections were rehydrated and incubated at 50°C for 30 min in hybridization buffer (50% deionized formamide, 4x SSC (0,6 M NaCl, 0,06 M sodium citrate; pH 7.2), 0,4 % sodium dodecyl sulfate (SDS), 0,2 % blocking reagent (Roche, Mannheim, Germany), Denhardt’s (Sigma-Aldrich, Steinheim, Germany), 0,2 mM ethylene-diamine tetra-acetic acid (EDTA) (pH 8), 0,04 mg/ml Salmon Sperm DNA Solution (Invitrogen, Carlsbad, CA, USA), 0,625 U/ml Transfer ribonucleic acid from baker's yeast (Sigma-Aldrich, Steinheim, Germany). Hybridizations were performed overnight at 42°C with a 1:5 probe dilution. The sections were washed twice for 10 min in 4x SSC, 0.1% SDS at room temperature and twice for 10 min in 0.1x SSC, 0.1% SDS at 50°C with agitation. Bound probes were detected by alkaline phosphatase immunostaining. Stained nematode sections were examined with differential interference contrast microscopy. References Altschul S, Gish W, Miller W, Myers E, Lipman D. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. de Boer JM, Yan Y, Smant G, Davis EL, Baum TJ. 1998. In-situ hybridization to messenger RNA in Heterodera glycines. Journal of Nematology 30: 309–312. Ewing B, Green P. 1998. Base-calling of automated sequencer traces ssing Phred.II error probabilities. Genome Research 8: 186–194. Ewing B, Hillier L, Wendl MC, Green P. 1998. Base-Calling of Automated Sequencer Traces Using Phred.I. Accuracy Assessment Genome Research 8: 175–185. Rice P, Longden I, Bleasby A. 2000. EMBOSS: The European molecular biology open software suite. Trends in Genetics 16: 276–277.