HW Answers pg. 241,2..
advertisement

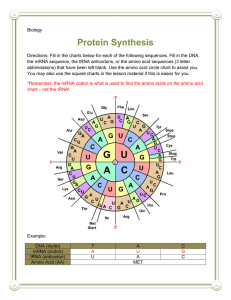
Answers to HW Questions Text Ques. Pg.241 • The DNA would be subjected to degradation, damage or distortion. The DNA is the original blueprint and must remain in the nucleus to be protected. • The central dogma of molecular genetics states that genes found within a DNA sequence are transcribed into m-RNA which then exits the nucleus & is translated into protein by ribosomes in the cytoplasm. • Ribosomes are the site of protein synthesis • M-RNA is formed by transcription and carries the message from the DNA to direct the protein being formed. • T-RNA contains the anti- codon and delivers the amino acids to the ribosome • R-RNA – helps to form & maintain ribosomes. Transcription Translation Location In the nucleus Purpose Copies a DNA sequence (a gene) into a template that can be used by the ribosome to build a protein. In the cytoplasm (ribosomes) Uses the mRNA template in order to build the specified protein. • A start codon (AUG) tells the ribosome where to begin the production of a polypeptide chain. • A stop codon tells the ribosome when a polypeptide chain ends. (UAA,UGA,UAG) • CCU AGU UCC AGG UCC GUU AAA UCG UAC GGG GUU 11. Using the genetic code, decipher the following mRNA sequence: 5 - GGCAUGGGACAUUAUUUUGCCCGUUGUGGUGGGGCGUGA - 3 • DNA sequence divided into codons • 5'–GGC-AUG-GGA-CAU-UAU-UUU-GCC-CGUUGU-GGU-GGG-GCG-UGA–3' • The start codon is the second codon, AUG; therefore, translation into protein commences with the amino acid methionine. • Translation results in the following protein: • Met-Gly-His-Tyr-Phe-Ala-Arg-Cys-Gly-Gly-Ala • The last codon, UGA, is a stop codon. 13. The amino acid sequence for a certain peptide is Leu–Tyr–Arg–Trp–Ser. How many nucleotides are necessary in the DNA to code for this peptide? • Since there are five amino acids found in this polypeptide, there must be at least five codons. Since each codon consists of three nucleotides, 15 (5×3) nucleotides would be required to code for this peptide sequence. Ques. Pg.249 1. Explain the role of the following in transcription: RNA polymerase, . poly-A polymerase, and spliceosomes • RNA polymerase plays many roles in the process of transcription. It recognizes the promoter region upstream of a gene to • be transcribed and binds to this site. Binding to the promoter region results in DNA’s double helix opening up. Once • bound to the DNA template strand, RNA polymerase starts to build the mRNA complementary strand using • ribonucleotides. Finally, RNA polymerase recognizes the termination sequence that signals the end of a gene and ceases • transcription. #1 cont’d • Poly-A polymerase plays a role in the posttranslational modification of mRNA. It adds approximately 200 to 300 adenine nucleotides to the 3' end of the primary mRNA transcript • Splicesomes found in eukaryotic organisms cut introns (noncoding regions) from the primary mRNA transcript and anneal the remaining exons (coding regions) together. 2.A short fragment of a particular gene includes the following sequence of nucleotides: 3- TACTACGGTAGGTATA - 5 Write out the mRNA sequence. • DNA 3'–TACTACGGTAGGTATA–5' • RNA 3'–AUGAUGCCAUCCAUAU–5' 3. A short fragment of a particular gene includes the following sequence of nucleotides: 3- GGCATGCACCATAATATCGACCTTCGGCACGG - 5 (a) Identify the promoter region. Justify your answer. (b) Explain the purpose of the promoter in transcription. • (a) 3' – GGCATGCACCATAATATCGAACCTTTCGGCA CGG – 5' • The red area represents the promoter region. The promoter region contains a high concentration of A and T bases. Since A and T only share two double bonds between them, RNA polymerase will expend less energy in opening up the double helix at this point. ( Known as the tata region) # 3 Cont’d • (b) The purpose of the promoter is twofold in the process of transcription. First, the promoter lies upstream of a DNA sequence that represents a gene. Hence, it acts as a signal for RNA polymerase to bind and transcribe the gene found downstream. Second, it also is an area where the DNA double strand is able to be unwound more easily because of its high concentration of adenine and thymine. 4. Differentiate between introns and exons. • Exons are segments of DNA that code for part of a specific protein, while introns are the noncoding regions of a gene. 5. In eukaryotes, mRNA must be modified before it is allowed to exit the nucleus. (a) Describe the modifications that are made to eukaryotic primary transcript. • The modifications that are made to the primary mRNA transcript include capping and tailing and the excision of introns. Capping involves the addition of a 7-methyl guanosine to the 5' end of the primary mRNA transcript. Tailing consists of the addition of 200 to 300 adenine nucleotides to the 3' end of the primary mRNA transcript by poly-A polymerase. The excision of introns is carried out by splicesomes, which then join the remaining exons together. 5.(b) Explain how these modifications ensure that mRNA survives in the cytoplasm and is translated into a functioning protein. • The capping and tailing of the primary mRNA transcript ensures that when the transcript exits the nucleus, it is not degraded by nucleases and phosphatases found in the cytoplasm. Capping also plays a role in the initiation of the process of translation. Introns are excised to ensure that when the mRNA transcript is translated into protein, it does not contain amino acids that are extraneous. These extraneous amino acids would interfere with the proper folding and functioning of the protein. 8. Explain the ramifications to the process of transcription if the following occurs: (a) The termination sequence of a gene is removed. (b) Poly-A polymerase is inactivated. • 8. (a) If the termination sequence of a gene is removed, RNA polymerase will continue to transcribe and build a primary mRNA transcript beyond the gene. • (b) If poly-A polymerase is inactivated, the 200 to 300 protective adenine nucleotides that are added at the end of a primary mRNA transcript will no longer be added. This will leave the mRNA susceptible to degradation on exiting the nucleus. 8. (c) The enzyme that adds the 5 cap is nonfunctional. (d) Spliceosomes excise exons and join the remaining introns together • (c) If the enzyme that adds the 5' cap is dysfunctional, the mRNA is susceptible to degradation on exiting the nucleus. • (d) If splicesomes excise exons and join introns together, then the mRNA transcript will consist of noncoding sequences.When the mRNA is translated, it will produce a nonfunctional protein. 8.(e) The RNA polymerase fails to recognize the promoter sequence. • (e) The RNA polymerase fails to recognize the promoter sequence. Ques. Pg. 254 1. Differentiate between the following: (a) P site and A site (b) codon and anticodon • (a) The A and P sites are found in a ribosome that is translating an mRNA sequence into protein. The A (acceptor) site is where tRNA molecules bring in the appropriate amino acid. The P (peptide) site is where peptide bonds are formed between adjoining amino acids on a growing polypeptide chain. • (b) A codon is a triplet of bases on mRNA that encodes a single amino acid. An anticodon is a triplet of bases on tRNA that recognizes and pairs with a codon on the mRNA. 3. List the possible anticodons for threonine, alanine, and proline. Amino Possible Acid codon ACU Threonine ACC Possible anti-codon ACA ACG UGA UGG UGU UGC Alanine GCU GCC GCA GCG CGA CGG CGU CGC Proline CCU CCC CCA CCG GGA GGG GGU GGC 4. Errors in the process of transcription are occasionally made. Explain why an error in the third base of a triplet may not necessarily result in a mutation. Provide an example to support your answer using the genetic code. • An error in the third base of a codon in mRNA may not necessarily result in an error during the process of translation because more than one codon encodes a particular amino acid. The codons differ by the third nucleotide. For example,proline can be encoded by the codons CCU, CCC, CCA, and CCG. If a mistake is made in the third nucleotide of the codon, it is negligible. It does not matter what the third nucleotide is—the two first nucleotides, CC, will always code for proline. 6. The following sequence was isolated from a fragment of mRNA: 5 - GGC CCA UAG AUG CCA CCG GGA AAA GAC UGA GCC CCG - 3 Translate the sequence into protein starting with the start codon. • The genetic code (Figure 7, Section 5.2, p. 240 of the Student Text) must be used. AUG is always the start codon and encodes for the amino acid methionine: • 5'–GGC-CCA-UAG-AUG-CCA-CCG-GGAAAA-GAC-UGA-GCC-CCG–3' • Met–Pro–Pro–Gly–Lys–Asp–Stop