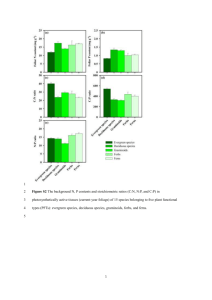
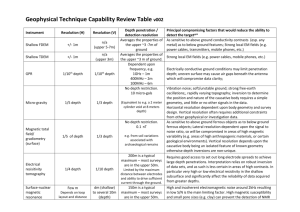
Legumes Graminoids Asters +
advertisement
Multiple comparisons - multiple pairwise tests - orthogonal contrasts - independent tests - labelling conventions Card example number 1 Multiple tests Problem: Because we examine the same data in multiple comparisons, the result of the first comparison affects our expectation of the next comparison. Multiple tests ANOVA shows at least one different, but which one(s)? •T-tests of all pairwise combinations significant significant Not significant Multiple tests T-test: <5% chance that this difference was a fluke… affects likelihood of finding a difference in this pair! Multiple tests Solution: Make alpha your overall “experiment-wise” error rate T-test: <5% chance that this difference was a fluke… affects likelihood (alpha) of finding a difference in this pair! Multiple tests Solution: Make alpha your overall “experiment-wise” error rate e.g. simple Bonferroni: Divide alpha by number of tests Alpha / 3 = 0.0167 Alpha / 3 = 0.0167 Alpha / 3 = 0.0167 Card example 2 Orthogonal contrasts Orthogonal = perpendicular = independent Contrast = comparison Example. We compare the growth of three types of plants: Legumes, graminoids, and asters. These 2 contrasts are orthogonal: 1. Legumes vs. non-legumes (graminoids, asters) 2. Graminoids vs. asters Trick for determining if contrasts are orthogonal: 1. In the first contrast, label all treatments in one group with “+” and all treatments in the other group with “-” (doesn’t matter which way round). Legumes + Graminoids - Asters - Trick for determining if contrasts are orthogonal: 1. In the first contrast, label all treatments in one group with “+” and all treatments in the other group with “-” (doesn’t matter which way round). 2. In each group composed of t treatments, put the number 1/t as the coefficient. If treatment not in contrast, give it the value “0”. Legumes +1 Graminoids - 1/2 Asters -1/2 Trick for determining if contrasts are orthogonal: 1. In the first contrast, label all treatments in one group with “+” and all treatments in the other group with “-” (doesn’t matter which way round). 2. In each group composed of t treatments, put the number 1/t as the coefficient. If treatment not in contrast, give it the value “0”. 3. Repeat for all other contrasts. Legumes +1 0 Graminoids - 1/2 +1 Asters -1/2 -1 Trick for determining if contrasts are orthogonal: 4. Multiply each column, then sum these products. Legumes +1 0 0 Graminoids - 1/2 +1 - 1/2 Asters -1/2 -1 +1/2 Sum of products = 0 Trick for determining if contrasts are orthogonal: 4. Multiply each column, then sum these products. 5. If this sum = 0 then the contrasts were orthogonal! Legumes +1 0 0 Graminoids - 1/2 +1 - 1/2 Asters -1/2 -1 +1/2 Sum of products = 0 What about these contrasts? 1. Monocots (graminoids) vs. dicots (legumes and asters). 2. Legumes vs. non-legumes Important! You need to assess orthogonality in each pairwise combination of contrasts. So if 4 contrasts: Contrast 1 and 2, 1 and 3, 1 and 4, 2 and 3, 2 and 4, 3 and 4. How do you program contrasts in JMP (etc.)? Treatment SS } Contrast 1 } Contrast 2 How do you program contrasts in JMP (etc.)? Normal treatments Legumes vs. nonlegumes Legume Legume Graminoid Graminoid Aster Aster 1 1 2 2 3 3 1 1 2 2 2 2 SStreat Df treat MStreat 122 2 60 67 1 MSerror Df error 10 20 “There was a significant treatment effect (F…). About 53% of the variation between treatments was due to differences between legumes and nonlegumes (F1,20 = 6.7).” F1,20 = (67)/1 = 6.7 10 From full model! Even different statistical tests may not be independent ! Example. We examined effects of fertilizer on growth of dandelions in a pasture using an ANOVA. We then repeated the test for growth of grass in the same plots. Problem? Multiple tests b a,b a significant Not significant Not significant Convention: Treatments with a common letter are not significantly different le ar d ed re d d un c ea cl un re l 30 Bi lik go ea cl un ua l bc Eb o ek ca ni an m ha e ab uf o og m ec pl et 20 Eb o o m 15 Eb og co w llo pl et e m co fa a Eb go ek m Mean no. butterflies/5 min.walk 10 Eb o uf Eb o Fa r 35 d 25 cd bc a 5 0