Slides
advertisement

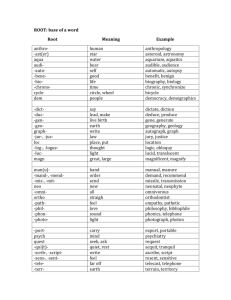
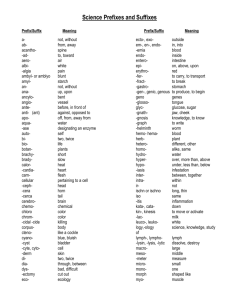
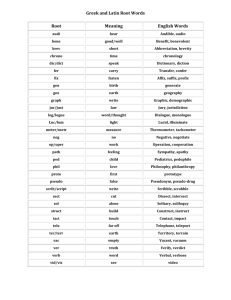
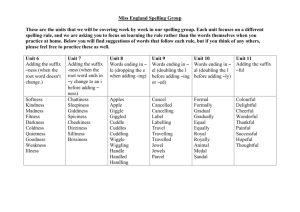
Database Index to Large Biological Sequences Ela Hunt, Malcolm P. Atkinson, and Robert W. Irving Proceedings of the 27th VLDB Conference,2001 Presented by Raghav & Balaji Indexing Large Biological Sequences Introduction Indexing strategies Suffix trees New Construction Algorithm Query Experiment and Results Conclusion Introduction What's a DNA? A, C, G, T (A with T, C with G) Base pair Gbp (Giga base pairs) Mammalian genomes – 3Gbp What is the challenge in indexing DNA? Large Size and no definite pattern Searching genetic DNA sequences Sequentially scanning and filtering approach (BLAST, FASTA) Introduction Rise in volume of data and demand for searches by researchers accelerated the need for better searches using indexes. New Sequences will be revealed as improved sequencing techniques are developed. Determining DNA sequences is useful in studying fundamental biological processes, as well as in forensic research. Indexing Strategies Considered Inverted files Not suitable since DNA cannot be broken into words. B-tree Same as above Q-grams Cannot deliver matches that have low similarity to the query. Most of the techniques are infeasible. Indexing Strategies Considered Suffix Trees Ideal Choice for this type of indexing. Suffix trees on disk could only be built for small sequences. “Memory Bottleneck”. Suffix tree storage optimization Reduce the RAM required to around 13 bytes per character indexed Not test on disk Indexing Strategies Considered Approach to searching genetic DNA sequences using an adaptation of the suffix tree. Build suffix tree on disk for arbitrarily large sequences New query process strategies. Alternative data structures Q-grams, Suffix array, String B tree… Suffix Trees Suffix tree - compressed digital trie. A suffix tree is a rooted directed tree with m leaves, where m is the length S (the database string) For any leaf i, the concatenation of the edgelabels on the path from the root to leaf i exactly spells out the suffix of S that starts at position i Suffix Trees Suffix tree is a compressed digital (suffix) trie Suffix tree building Suffices of mississippi: 1 mississippi 2 ississippi 3 ssissippi 4 sissippi 5 issippi 6 ssippi 7 sippi 8 ippi 9 ppi 10 pi 11 i root i s s i s s i p p i i Result suffix tree building p root s i i 9 11 i ssi 10 si 8 4 1 2 5 6 3 7 Suffix Trees Suffix Links: A necessary implementation trick to achieve a linear time and space bound during building the tree A suffix link is: a pointer from an internal node xS to another internal node S where x is a arbitrary character and S is a possibly empty substring Suffix Trees Construction Suffix link Complexity O(n) Ukkonen’s Method Suffix Trees General applications of Suffix trees Find all occurrences of q as a substring of S Longest substring common to a set T of strings Find the longest palindrome in S Suffix Trees Analysis of Suffix Link Based Algorithm Build the tree incrementally, check pointing the tree after each portion has been attempted. 2 distinct traversal patterns exist both of which are used during construction. Very long construction time. These effects combine to limit the size of the tree that can be constructed and stored on disk to the available main memory. Suffix Trees Using Suffix link based algorithm, it was observed that checkpointing trees indexing more than 21Mbp was not possible using 1.8GB of main memory. Reasons being Object header size increases New Construction Algorithm Difficulties of traditional suffix tree construction: Memory bottleneck Necessity of random access New conception To abandon the use of suffix links To perform multiple passes over the sequence, constructing the suffix tree for a sub range of suffixes at each pass. New Construction Algorithm Removing Suffix link means that the construction of a new partition does not modify previously checkpointed partitions of the tree. Using multiple passes, it means that it is not necessary to access or update previously checkpointed partitions. i.e. Data structure for the complete partitions can be evicted from the main memory and will not be faulted back during the rest of the tree’s construction. New Construction Algorithm Partition concept: Build multiple suffix tree that fit in memory(AC, AT or AG fall into different partitions) Base on the prefixes of each suffix Use a sliding window of length l. Form a string s1 of window length, l. Scan the string and count the number of occurrances of s1. Use a bin packing technique to pack (s1, #occurrances) New Construction Algorithm Partition technology: Assumption:tree is uniformly populated. Prefix code(Pi): Suffixes that are indexed during the jth pass of the sequence have jr Pi (j+1)r New Construction Algorithm The actual algorithm [Pseudo code] New Construction Algorithm 1 2 3 4 5 root ANA$ NA$ A$ $ Tree creation for ANA$ root $ ANA$ A NA$ 2 NA$ 2 3 $ 4 5 New Construction Algorithm Original tree (Ukkonen) left index right index suffix number Modified Node left index child child sib suffix link sib Query Only exact pattern matching. One query involves one partial traversal. Complexity of suffix tree search: O(k+m); k-query length, m-no of matches in the index. Queries of length q bring back 1/(a^q) fraction of the whole tree where a = size of the active alphabet i.e. 4 (A,C,G,T). New query strategies: Short query: serial scan of the sequence Longer query: using index structure Threshold: 10 to 12 letters Experiment and Results Develop and experiment platform: Software: PJama, JAVA 1.3 & Solaris 7 OS Hardware: Enterprise 450 with 2GB RAM Test data 6 single chromosomes of worm C. elegans(20.5Mbp max. length) Human chromosomes 21,22, and 1(280Mbp) Alphabets A, C, G, T, $, * Experiment and Results Trees with suffix link: (use 20.5Mbp DNA) – Construct in memory: 7 mins – Construct in disk: 34 hours Trees without suffix link: (263Mbp DNA) – 19 hours Experiment Results Exact String matching using 263Mbp of human DNA Queries sent in batches using warm storage Experiment Results Cold Storage Experiment Results Further Work Improvements to the tree representation and incremental construction algorithm. Investigation of the interaction between approximate matching algorithms and diskbased suffix trees. Investigation of alternative persistent storage solutions. Integration of the algorithms with biological research tools and usability studies. Conclusion Present an approach to searching genetic DNA sequences using an adaptation of the suffix tree data structure. Allow to build suffix trees on disk for arbitrarily large sequences. Open up the perspective of building suffix trees in parallel, and the simplicity of this approach can make suffix trees more popular.