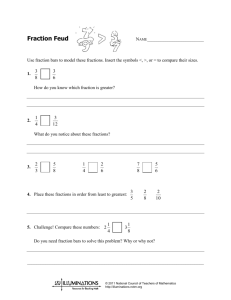
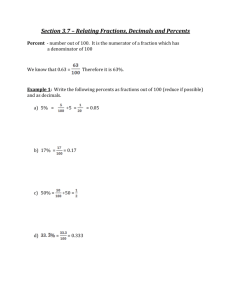
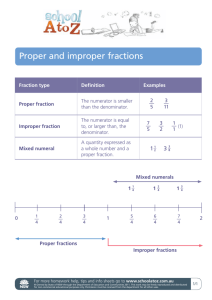
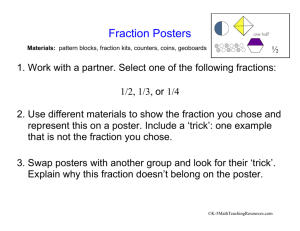
Supplementary Materials
advertisement

Fig. S1: LPS profiles and membrane fractions. (a) LPS profiles of wild type, dnpA mutant and dnpA overexpressing strains. LPS was isolated and analyzed as described in material and methods. LPS profiles of WT and dnpA mutant (dnpA-) (left panel) and of WT containing plasmid pME6032 (WT\pME6032) and WT overexpressing dnpA from the plasmid pME6032 (WT\pME6032-dnpAHis) (right panel). (b) SDS-PAGE analysis of outer (OM) and inner membrane (IM) fractions of wild type and dnpA mutant (dnpA-) respectively. Membrane fractions were isolated as described in material and methods. M: Wide range SigmaMarker (SigmaAldrich). Fig. S2: Western blot analysis of cells expressing wild-type and mutant dnpA alleles from pME60332. Precision Plus Protein Kaleidoscope Standards (Bio-Rad) was used as molecular marker. The molecular weight of DnpA-His is approximately 54,64 kDa. Fig. S3: Evolution of the persister fraction after dilution into fresh medium. Stationary phase cells were diluted 100-fold into fresh medium. Every hour, an aliquot was taken of which A. the optical density (595 m) and B. the persister fraction was monitored. The persister fraction was determined following a 5 h treatment with ofloxacin. Untreated cells were used as a control. Data points correspond to the mean relative persister fraction. Each experiment was repeated independently at least 3 times. Error bars represent the standard error of the mean (SEM). A relative persister fraction of 100 % corresponds 2.15 × 10-5. Samples for RNA isolation were taken at an OD595 of 1 when the persister fraction started to increase (indicated by an arrow). Table S1: List of genes differentially expressed in dnpA mutant and overexpressing strain (FDR 15 %).