Course 7: LC-MS data handling - Metabolomics Fiehn Lab
advertisement

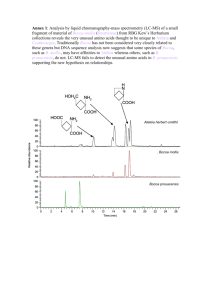
Welcome! Mass Spectrometry meets Cheminformatics Tobias Kind and Julie Leary UC Davis Course 7: Concepts for LC-MS Class website: CHE 241 - Spring 2008 - CRN 16583 Slides: http://fiehnlab.ucdavis.edu/staff/kind/Teaching/ PPT is hyperlinked – please change to Slide Show Mode General LC-MS data processing for small molecules Confirm with MS/MS or MSn fragmentation 100 50 MS2 0 200 400 600 Deconvolution and evaluation of LC-MS data UPLC_C8_DataDependent_Chlamy_07030703... 3/7/2007 3:21:28 AM RT: 0.00 - 44.99 100 17.80 90 NL: 1.68E6 18.30 m/z= 70.00000-2000.00000 F: FTMS + p NSI Full ms [200.00-1200.00] MS UPLC_C8_DataDependent _Chlamy_070307032128 19.11 LC-MS Chromatogram 80 70 Picture: nrel.gov 60 29.36 LC-MS run 40 minutes C8 column, Agilent-UPLC Chlamydomonas extract 28.74 50 20.16 40 30.30 11.27 43.97 30.55 23.00 30 17.49 43.68 23.80 20 10.31 10 1.56 6.37 16.07 9.55 31.53 43.09 37.19 0 0 5 10 15 20 25 Time (min) 30 35 40 + UPLC_C8_DataDependent_Chlamy_070307032128 #1414 RT: 18.30 AV: 1 NL: 8.23E5 F: FTMS + p NSI Full ms [200.00-1200.00] 756.57745 R=55401 100 90 80 Extracted Mass spectrum 70 60 757.58209 R=55100 50 40 30 20 756.26086 R=87000 10 754.57471 R=50200 0 754 755 755.57501 R=60200 756 758.58636 R=56100 756.89417 R=86600 757 759.59546 R=60400 757.90027 R=79200 758 759 m/z Chromatogram Source: N.Saad, DY Lee FiehnLab 760 760.60144 761.59888 762.29700 R=52200 R=62600 R=49901 761 762 FT-ICR-MS mass spectrum MS1 @ 50,000 resolving power Check charge state = 1 756.57 represents [M+H]+ Peak Picking with ACD/SpecManager 9.0 Processing of LC-MS data - use of MassFrontier Deconvolution and evaluation of LC-MS data Example with HighChem Mass Frontier LC-MS detected compound Marked with blue triangle LC-MS detected compound Marked with blue triangle 141 peaks extracted Extracted MS1 peak Library search useless (only single peak) Adduct removal and detection during ESI-LC-MS runs 100 756.57745 R=55401 [M+H]+ = 756.577 M = 755.5627 90 80 Relative Abundance 70 60 757.58209 R=55100 50 40 30 20 758.58636 R=56100 10 759.59546 R=60400 0 756.5 Ion name 757.0 757.5 Ion mass 758.0 m/z 758.5 759.0 759.5 Your M here: 755.562724 Your M+X or M-X 756.577 Charge Mult Mass Result: Reverse: 1. Positive ion mode M+2H M/2 + 1.007276 M+H+NH4 M/2 + 9.520550 M+H+Na M/2 + 11.998247 M+H M + 1.007276 M+NH4 M + 18.033823 M+Na M + 22.989218 2+ 2+ 2+ 1+ 1+ 1+ 0.5 0.5 0.5 1 1 1 1.007276 9.520550 11.998247 1.007276 18.033823 22.989218 378.788638 387.301912 389.779609 756.570000 773.596547 778.551942 377.277724 368.764450 366.286753 755.562724 738.536177 733.580782 2. Negative ion mode M-3H M/3 - 1.007276 M-2H M/2 - 1.007276 M-H2O-H M- 19.01839 M-H M - 1.007276 M+Na-2H M + 20.974666 M+Cl M + 34.969402 321111- 0.33 0.5 1 1 1 1 -1.007276 -1.007276 -19.01839 -1.007276 20.974666 34.969402 250.846965 376.774086 736.544334 754.555448 776.537390 790.532126 253.197276 379.292276 775.588390 757.577276 735.595334 721.600598 Download Adduct-Calculator [LINK] Problem: Detection of molecular ion Problem: Is this the pure mass spectrum or from overlapping peaks? Is it M+H or M+Na or any of the 40 other adducts? Example data from crocin standard mixture (expected MW: 976.965) 27-2-crocin-LTQFT-pos-0V #1 RT: 0.00 AV: 1 T: FTMS + p NSI Full ms [ 50.00-2000.00] 27-2-crocin-LTQFT-pos-100V #1 RT: 0.01 AV: 1 NL: 1.30E4 T: FTMS + p NSI sid=100.00 Full ms [ 50.00-1500.00] NL: 9.78E3 719.21790 100 [M+Na]+ 95 95 90 999.37189 100 90 CID = 0V 85 85 80 80 75 75 70 70 65 65 60 60 CID = 100V Relative Abundance 719.21747 55 50 45 55 749.22906 50 45 529.17432 40 40 1035.36377 367.12146 575.21149 35 35 30 25 30 [M+Na]+ 853.28149 1015.33575 675.26556 529.17535 25 853.28583 20 20 367.12228 15 575.21246 1177.38086 301.14145 1241.42676 10 10 1359.45837 1517.50293 5 1695.55408 400 600 800 1000 m/z 1200 1400 1600 329.95432 219.18610 5 0 200 1177.39673 15 185.11496 1800 485.20132 969.36615 77.14542 0 2000 200 400 600 800 m/z 1000 1225.41028 1339.45142 1200 1400 Adduct removal and detection during LC-MS runs Ion name Ion mass 1. Positive ion mode M+2H M/2 + 1.007276 M+H+NH4 M/2 + 9.520550 M+H+Na M/2 + 11.998247 M+H M + 1.007276 M+NH4 M + 18.033823 M+Na M + 22.989218 2. Negative ion mode M-3H M/3 - 1.007276 M-2H M/2 - 1.007276 M-H2O-H M- 19.01839 M-H M - 1.007276 M+Na-2H M + 20.974666 M+Cl M + 34.969402 Charge Mult Mass Your M here: 853.33089 Your M+X or M-X 756.577 Result: Reverse: 2+ 2+ 2+ 1+ 1+ 1+ 0.5 0.5 0.5 1 1 1 1.007276 9.520550 11.998247 1.007276 18.033823 22.989218 427.672721 436.185995 438.663692 854.338166 871.364713 876.320108 437.152724 428.639450 426.161753 875.312724 858.286177 853.330782 321111- 0.33 0.5 1 1 1 1 -1.007276 -1.007276 -19.01839 -1.007276 20.974666 34.969402 251.182724 377.277724 737.551610 755.562724 777.544666 791.539402 253.197276 379.292276 775.588390 757.577276 735.595334 721.600598 Adduct can be removed before or after formula generation. For good isotopic pattern matching remove adduct after formula generation. 7 Golden Rules apply LEWIS and SENIOR check (adduct needs to be removed) Formula Generation from accurate mass measurement CHN5O13P15 or CHN11P19 ??? Apply Seven Golden Rules for correct molecular formulas Apply heuristic and mathematical and chemometric rules for filtering elemental compositions Isotopic Pattern generator from formula Example from MWTWIN Is usually included in every LC-MS software Isotopic pattern equally important as accurate mass Experimental result Abundances for all molecular formulae 25.00 756.57745 R=55401 100 90 20.00 Isotopic Abundance A+2 [%] 80 A+1 = 47.44% Relative Abundance 70 60 757.58209 R=55100 50 40 A+2 = 10.92 % 30 20 758.58636 R=56100 10 15.00 10.00 5.00 759.59546 R=60400 0 756.5 757.0 757.5 758.0 m/z 758.5 759.0 759.5 0.00 0.00 20.00 40.00 60.00 80.00 Isotopic Abundance A+1 [% ] m/z Intensity Relative Resolution 756.5765 757.5814 758.5862 756.5526 759.593 589034.1 279455.2 64293.3 19173.8 9656.4 100 47.44 10.92 3.26 1.64 54901 55100 56100 52000 56000 We can discard all other results outside the error box. Current box reflect +/- 10% error. Problems during LC-MS peak detection and MS deconvolution 48 Multiple peaks detected But it is a single component only 36 24 12 0 10.80 10.90 11.00 11.10 11.20 11.30 11.40 11.50 11.60 11.70 11.80 11.90 Multiple peaks detected – Solution: adjust deconvolution settings Mass spectra not clean – Solution: manual peak extraction Not enough peaks detected – Solution: increase signal noise (S/N) settings Finding optimum settings is: • non-trivial and can change in different matrices • can be evaluated on standards and quality check mixtures • can be obtained by self-sharpening algorithms UPLC-FT-MS data extraction with MassFrontier Mass Frontier 5.0 Report File MS1 FTMS + p NSI Full #1414 MS1 100 Fragment peaks m/z 478.45 and 496.46 756.577 50 0 755 100 760 478.45 496.46 75 MS2 File MS 2 ITMS + c NSI d Full 756.58 #1415 100 MS2 50 478.45 496.46 50 25 335.56 0 200 400 600 0 236.20 271.06 250 300 335.56 350 391.34 434.52 400 450 500 550 600 650 687.72 700 100 756.577 75 MS1 50 25 0 751 752 753 754 755 756 757 758 759 760 761 762 Approach: generate molecular formula using Seven Golden Rules; find matching isomers in molecular databases; confirm possible matches by in-silico fragmentation (usually impossible); Seven Golden Rules – generate possible molecular formulas 5 formula candidates left with 30 ppm mass accuracy and 10% isotopic abundances These are candidates with good isotopic pattern match. These 5 were found in PubChem. C42H78NO8P - 1 isomer hit C42H77NO10 - 1 isomer hit C39H73N5O9 - 0 isomer hit C43H82NO7P - 2 isomers found C43H73N5O6 -2 isomers found C45H77N3O6 - 1 hit found C45H69N7O3 - 1 hit found Scan speed problem: Due to poor ion statistics only few scans are collected Mass accuracy and isotopic abundance accuracy are bad Structural isomer lookup example in ChemSpider In-silico fragmentation with MassFrontier using fragmentation library of 20,000 mechanisms from literature In-silico fragmentation with MassFrontier Experimental peaks m/z 478.45 and 496.46 were detected in MS/MS spectrum In-silico fragmentation should match the experimental fragmentation. In-silico - using a computer library of 20,000 fragmentation rules from the MS literature C42H78NO8P – Fragments N.A. C42H77NO10 – Fragments N.A. C43H82NO7P – Fragments m/z 478 and 496 LWHKEEJOPDYARM-WYCAKVPNBV Possible solution (2 fragments match) C45H69N7O3 – Fragments: m/z 496 C43H82NO7PH – Fragments N.A. Discussion of general LC-MS approach Example discussion: Fragment at m/z 236 not explained; molecular ion may be wrong; Substance can be potential new compound Must be confirmed by NMR or external standard General problems: Best approach is to generate MS and MSn and MSe mass spectral libraries Adduct removal is a problem Building target lists is always good (know what to expect) Focus on certain substance classes only Focus on single compound only Substance must be known for in-silico approach Fragmentation rules must be captured for in-silico approach In-silico approaches work best for peptides, carbohydrates, lipids (due to known and stable fragmentations) The Last Page - What is important to remember: Always use peak picking and mass spectral deconvolution for LC-MS data Apply accurate mass, accurate isotopic abundances together for formula generation Make use of high resolving power whenever possible Use MS/MS data and mass spectra from different ionization voltages Use existing MS/MS libraries or create your own MSn tree libraries Use molecular isomer databases for obtaining possible structure candidates Confirm if possible with MSn data or other possible filter constraints Tasks (36 min): (1) Download and install one tool from http://www.ms-utils.org/wiki/pmwiki.php/Main/SoftwareList (2) Download sample data from Open Data repositories and report your experience with the selected software (1) on the sample data Reading List (44 min): Hardware and Software Challenges for the Near Future: Structure Elucidation Concepts via Hyphenated Chromatographic Techniques QUALITATIVE AND QUANTITATIVE MASS SPECTRAL ANALYSIS Congruent Strategies for Carbohydrate Sequencing. 3. OSCAR: An Algorithm for Assigning Oligosaccharide Topology from MSn Data Data Reduction of Isotope-resolved LC-MS Spectra Comparative LC-MS: A landscape of peaks and valleys Links: Used for research: (right click – open hyperlink) Mass Spectrometry in Life Science: Technology and Data-Evaluation Managing and Exploring Large Data Sets Generated by Liquid Separation - Mass Spectrometry Metabolomics Technologies applied to the Identification of Compounds in Plants Of general importance for this course: http://fiehnlab.ucdavis.edu/staff/kind/Metabolomics/Structure_Elucidation/