Genetics --- introduction
advertisement

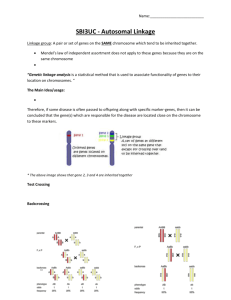
B2250 Readings and Problems ***Note: Quiz #2: Thurs. Oct. 28 Ch. 4 p. 100 – 112 Ch. 5 p. 118 – 137 Ch. 6 p. 148 – 165 Prob: 10, 11, 12, 18, 19 Prob: 1 – 11, 13 Prob: 1-5, 7, 8, 10, 11, 14 Practice problems: http://www.mun.ca/biology/dinnes/B2250/B2250.html (also download Lab. #3 and Print out 3 files) Genetics in the news “Junk DNA??” http://www.jgi.doe.gov/science/highlights/nobrega 1004.html Mendelian Genetics Topics: -Transmission of DNA during cell division Mitosis and Meiosis - Segregation - Sex linkage (problem: how to get a white-eyed female) - Inheritance and probability - Mendelian genetics in humans - Independent Assortment - Linkage - Gene mapping - 3 point test cross - Tetrad Analysis (mapping in fungi) - Extensions to Mendelian Genetics - Gene mutation - Chromosome mutation - Quantitative and population genetics Two Characters Monohybrid Cross parents differ for a single character (single gene ); seed shape Dihybrid Cross parents differ for two characteristics (two genes) Dihybrid Two Characters: 1. Seed colour yellow green Y y 2. Seed shape Round wrinkled R r 4 phenotypes Dihybrid P RRyy Gametes F1 X Ry rrYY rY RrYy DIHYBRID F1 Dihybrid ----->F2 F1 RrYy RrYy F2 9 3 3 1 Total 315 108 101 32 556 X RrYy round, yellow round, green wrinkled, yellow wrinkled, green Individual Characters 1. Seed shape round : wrinkled 423 : 133 3: 1 (¾ : ¼) 2. Seed colour yellow : green 416 : 140 3 : 1 Conclusion * 3 : 1 monohybrid ratio for each character * 9 : 3 : 3 : 1 phenotypic ratio a random combination of 2 independent 3:1 ratios Two Independent Genes F2 colour yellow 3/4 green 1/4 seed shape 3/4 1/4 round wrinkled 9/16 3/16 3/16 F2 1/16 Phenotypes Independent Assortment Test Cross AaBb X gametes ab 1/4 AB AaBb 1/4 Ab Aabb 4 gamete types 1/4 aB aaBb 1/4 ab aabb aabb 4 phenotypes 4 genotypes (Genes) Meiosis I A Correlation of genes and Chromosomes during meiosis a 4 gamete types A B A b OR a b a B Mendel’s Second Law Independent assortment: during gamete formation, the segregation of one gene pair is independent of other gene pairs. Linkage Chapter 6 - recombination - linkage maps Linkage of Genes - Many more genes than chromosomes - Some genes must be linked on the same chromosome; therefore not independent Independent Assortment F1 AaBb X AaBb Genotypes AABB AaBb AaBB F2 9 3 4 phenotypes 3 1 A-BA-bb aaBaabb AABb Aabb, AAbb aaBb, aaBB Independent Assortment Test Cross AaBb X gametes ab 1/4 AB AaBb 1/4 Ab Aabb 1/4 aB aaBb 1/4 ab aabb aabb 4 phenotypes 4 genotypes Independent Assortment Fig 6-6 AB ab Ab aB Interchromosomal Recombination Complete Linkage P X A F1 F1 gametes B a b A B a b AaBb A B AB dihybrid Parental Parental a b ab Recombinant Gametes ? Crossing over: - exchange between homologous chromosomes Crossing over in meiosis I Meiosis I - homologous chromosomes pair - reciprocal exchange between non-sister chromatids Crossing over in meiosis I (animation) Gamete Types F1 gametes A B a b A a A a B b b B AaBb AB ab Ab aB Parental Parental Recomb. Recomb. 1. Ways to produce dihybrid P Cis A B A B X Note: Chromatids omitted a b a b A B a b Gametes: AB ab Ab aB AaBb (dihybrid ) P P R R 2. Ways to produce dihybrid a B X a B AaBb A b trans (dihybrid ) a B Gametes: P Ab P aB R AB R ab P A b A b Two ways to produce dihybrid A B a b X A B a b cis A B a b Gametes: AB ab Ab aB P AaBb (dihybrid ) P P R R A b A b a B X a B A b trans a B Ab aB AB ab Independent Assortment Fig 6-6 Interchromosomal Linkage Fig 6-11 Intrachromosomal Example Test Cross How to distinguish: Parental high freq. Recombinant low freq. AaBb AB Ab aB ab X ab AaBb Aabb aaBb aabb aabb Exp. 25 25 25 25 100 Obs. 10 R 40 P 40 P 10 R 100 Example (cont.) Gametes: AB R Ab P aB P ab R Therefore dihybrid: A a b (trans) B Linkage Maps Genes close together on same chromosome: - smaller chance of crossovers between them - fewer recombinants Therefore: percentage recombination can be used to generate a linkage map Linkage maps A a C c B b D d large # of recomb. small number of recombinants Linkage maps example Testcross progeny: P AaBb 2146 R Aabb 43 65 R aaBb 22 4513 = 1.4 % RF P aabb 2302 Total 4513 1.4 map units A 1.4 mu B Additivity of map distances separate maps A B A 7 combine maps C 2 A 2 B 7 or A C 2 C B 5 Locus (pl. loci) Summary Mendelian Genetics: Monohybrid cross (segregation): Dihybrid Cross (Indep. Assort.): - ratios (3:1, 1:2:1, 1:1) - ratios (9:3:3:1, 1:1:1:1) - dominance, recessive - linkage (deviation from I.A.) - autosomal, sex-linked - recombination - probability - linkage maps - pedigrees Linkage Deviations from independent assortment Dihybrid gametes 2 parent (noncrossover) common 2 recombinant (crossover) rare % recombinants a function of distance between genes % RF = map distance Gametes Number of Genes Number of Different Gametes monohybrid 1 (Aa) 2 dihybrid 2 (AaBb) 4 trihybrid 3 (AaBbCc) ? Three Point Test Cross Trihybrid AaBbCc ABC ABc AbC Abc aBC aBc abC abc X aabbcc abc 8 gamete types Three Point Test Cross Trihybrid Gametes C ABC c ABc C AbC c Abc B A b a Three Point Test Cross Trihybrid AaBbCc 3 genes: Possibilities: 1. All unlinked 2. Two linked; one unlinked 3. Three linked Three Point Test Cross Three recessive mutants of Drosophila: P +/+ cv/cv ct/ct +, v vermilion eyes +, cv crossveinless +, ct cut wing X v/v +/+ +/+ Three Point Test Cross P +/+ cv/cv ct/ct Gametes F1 trihybrid + cv ct x v/v +/+ +/+ v + + v/+ cv/+ ct/+ Three Point Test Cross F1 v/+ cv/+ ct/+ 8 gamete types x v/v cv/cv ct/ct v cv ct one gamete type 8 gamete types F1 v/+ cv/+ ct/+ v + + cv v cv + + v cv + + v + + cv + 580 ct 592 + 45 ct 40 ct 89 + 94 ct 3 + 5 1448 Parental Parental Recombinant 8 gamete types F1 v/+ cv/+ ct/+ v + + cv v cv + + v cv + + v + + cv + 580 ct 592 + 45 ct 40 ct 89 + 94 ct 3 + 5 1448 Parental Recombinant Recombinant Parental 8 gamete types F1 v/+ cv/+ ct/+ v + + cv v cv + + v cv + + v + + cv + 580 ct 592 + 45 ct 40 ct 89 + 94 ct 3 + 5 1448 Parental Parental Recombinant Recombinant 8 gamete types F1 v/+ cv/+ ct/+ v + + cv v cv + + v cv + + v + + cv + 580 ct 592 + 45 ct 40 ct 89 + 94 ct 3 + 5 1448 Parental Recombinant Parental Recombinant Calculate Recombination Fraction 1. v - cv 2. v - ct 3. ct - cv R v cv R + + R + + R v ct R ct + R + cv 45 + 89 40 + 94 268 / 1448 = 18.5 % 94 + 5 89 + 3 191/1448 = 13.2 % 40 + 3 45 + 5 93/1448 = 6.4 % Three point test cross Observations: all 3 RF < 50 % 3 genes on same chromosome v-----cv largest distance ct in middle map v-------ct-------cv = cv-------ct-------v 13.2 + 6.4 = 19.6 > 18.5 !! Why ? Three Point Test Cross P +/+ ct/ct cv/cv gametes F1 trihybrid + ct x v/v +/+ +/+ cv v + v + + ct + cv + Three Point Test Cross Double crossover class rarest: v---cv P v P + R R v + X + ct ct + X + cv v + + cv + cv v + + cv Three Point test cross 1. Double crossovers not counted in v--cv RF 2. Double crossovers generate P types (with respect to v--cv) 3. Double crossovers not detected as recombinants Consequence: underestimate of v----cv map distance Greater distance of genes greater error Double recombinant class: (3 + 5) x 2 = 16 268 + 16 = 284 284/1448 = 19.6 NOTE: double crossovers detected because of middle gene (ct) Q. Are multiple crossovers independent ? Example: v 13.2 ct 6.4 cv Prob.(single recombinant v---ct) = 0.132 Prob.(single recombinant ct---cv) = 0.064 If independent then: Prob. (double recombinant)= 0.132 x 0.064 = 0.0084 expected number of doubles = 1448 x 0.0084 = 12 Double Cross Overs Expect 12 Observe 8 Explanation: a crossover in one region reduces the probability of a second crossover in an adjacent region Interference Interference (I) 1. Coefficient of coincidence (CC) Obs # double recombinants CC = Exp # double recombinants 2. Interference: I = 1 - CC = 1 - (8 /12) = 1/3 = 33 % I = 1 interference complete I = 0 no interference Linkage Other Points: 1. No crossing over in male Drosophila male: AaBb A B gametes AB, ab a b use female dihybrid: AaBb x aabb O O Linkage 2. Linkage of genes on the X chromosome: AaBb x --Y O O Male progeny: AB Y Ab Y male progeny direct aB Y measure of female meiotic ab Y products Mapping Function Genes close together on chromosome -RF good estimate of map distance Genes far apart on chromosome - RF underestimates true map distance due to undetected multiple crossovers Mapping Function m = avg. # crossovers per meiosis (linear with true map distance) if m = 1 (1 cross over for every meiosis) then 50 % recombinants produced Therefore: map units (mu) = m x 50 Mapping Function Mapping function: - relates RF to true map distance (better estimate for genes separated by large distances) m = -ln (1 - 2RF) mu = m x 50 Mapping function Mapping Function Observed RF (%) 60 50 40 30 m = -ln(1 - 2RF) 20 10 0 0 50 100 150 True Map Distance (m x 50) 200 Mapping Function example 1. RF = 18.5 % m = 0.46 mu = 23.1 2. RF = 6.4 % m = 0.137 mu = 6.8 Summary: - short distances: use RF - long distances: use mapping function Fungal Genetics Fungi: important organisms in the ecosystem - decomposers - pathogens important for humans - food - pathogens (Biology 4040 – Mycology) Fun Facts About Fungi http://www.herbarium.usu.edu/fungi/funfacts/factindx.htm Fungi Neurospora crassa (bread mold) Morphological mutants Biochemical mutants (one gene, one enzyme) Linkage Map Neurospora crassa Linkage group I Fungus Life Cycle dominant stage haploid +, - mating types brief diploid stage meiosis n n + spores + meiosis n - 2n n Gamete Pool Gametes: Products of many meioses all pooled together A B a b AB AB ab ab AB ab P AB ab ab AB ab Ab AB Gamete P ab AB aB ab ab AB AB pool R aB ab AB AB ab R Ab Tetrad Analysis Some Fungi and algae: 4 products of a single meiosis can be recovered Advantages: 1. haploid organism - no dominance 2. examine a single meiosis - test cross not needed 3. small, easy to culture 4. Tetrad Analysis - map gene to centromere Ascus with ascospores Tetrad Analysis Types of Tetrads: 1. Unordered - 4 products mixed together 2. Ordered (linear) - 4 products lined up, each haploid nucleus can be traced back through meiosis 3. Octads - mitotic division after meiosis 8 products (2 x 4) Figure 6-5 Linear Tetrad Analysis Life Cycle: + = a+ a a a + + a + a Meiosis + Diploid Haploid Mating: a n + x + a + n 2n 4 haploid products Linear Tetrad Analysis a a a a + + + + 8 haploid spores mitosis a a + + 4 haploid products (Octad) Linear Tetrad Analysis Two types of asci: 1. no crossover----> first division segregation (MI) 2. crossover between gene and centromere-----> second division segregation (MII) Mapping gene to centromere First Division a a + + No Crossover a a a a + + + + First division segregation A A A A a a a a Mapping gene to centromere Second division a a a + a + + a + a + + crossover Second division segregation A A a a A A a a Mapping gene to centromere I a a a a + + + + 43 + + + + a a a a 43 II a a + + a a + + 3 + + a a + + a a 4 + a + a MI = 86 a + a + MII = 14 a + a + + a + a 3 4 Total = 100 Mapping gene to centromere MI = 86 MII = 14 14/100 = 14 % of meioses showed a crossover ½ of the crossover products recombinant RF = ½ x 14 % = 7 % a 7 m.u. Unordered Tetrad Analysis 1. still products of a single meiosis 2. can not map gene to centromere 3. linear tetrads can be analyzed as unordered 4. map distance between linked genes a b n X + + a b + + n 2n meiosis Unordered Tetrads Three kinds of unordered tetrads: a b ab a b+ + + ab a b+ meiosis a+b+ a+b +b+ +b a a 1. Parental Ditype 2. Nonparental Ditype 3. Tetratype PD NPD ab a b+ a+b a+b+ T PD ab ab a + b+ a + b+ T NPD ab a b+ a b+ a b+ a + b+ a+ b a+ b a+ b Unlinked genes PD = NPD a b X + + ab ab a b + + a/+, b/+ meiosis a + + b PD + + + + a + a + + b + b NPD Unordered Tetrads Unlinked Genes: PD = NPD Linked Genes: PD >> NPD NPD----> all products recombinant T--------> ½ products recombinant PD-----> all parental type PD = 58 RF = ½ T + NPD T = 40 T + NPD + PD NPD = 2 RF = 0.22 22 m.u. Tetrad Analysis Types of Tetrads: 1. Ordered (linear): map gene to centromere 2. Unordered: map genes Linkage: Summary • Recombination: generates variation (inter and intrachromosomal) • Genetic maps: - genes linked on the same chromosome - location of new genes relative to genes already mapped