side2
advertisement

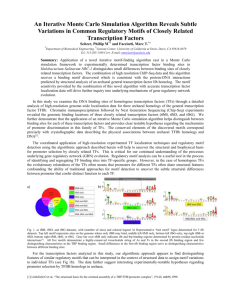
Distribution of DBDs in the TF Regulatory Network + + _ + _ _ _ metR _ marA rhaS marR + fadR iciA _ iclR uidR metJ _ _ dnaA _ rhaR uxuR soxS rob exuR lrp _ soxR mlc + + yiaJ malT melR araC nagC + malI glpR glnG fecI + + _ + + _ fucR + + + + gutM fur srlR _ + _ adiY rcsB + ompR csdG + _ + hns _ caiF tdcA _ mtlR arcA + betI lctR pdhR galR tdcR fnr Gene Product galS _ lysR “winged helix” DNA binding domain _ fis himA Homeodomain-like CRP fruR narL _ cspA _ Met repressor-like Nucleic acid-binding proteins Negative regulation with binding site data appY Putative DNA-binding domain C-terminal effector domain of the Bipartite response regulator IHF like DNA-binding domain Lambda repressor-like DNA binding domain Positive regulation with binding site data _ Negative regulation without binding site data + FIS-like Positive regulation without binding site data Transcription Factors and their regulated TFs Transcription Factors and their regulated genes A relatively complex topology exists in a simple organism The TF network has four major regulatory hubs which are also global regulators in their respective functional class. Information on the regulated gene is available for 113 TFs from RegulonDB. We classified the 113 TFs in to nine functional classes according to what they respond to and what they regulate. Each of these functional classes have one to three proteins (global regulators) that regulate over 50 genes and the other proteins (fine tuners) control fewer than 20 genes Number of Examples 18 16 14 12 10 8 6 4 2 0 X=1 X=2 X=3 X=4 X = 17 Number of different TF genes (X) regulated by a transcription factor Very few TFs regulate more than one TF Network Growth by Gene Duplication Duplication growth models Model-1: Duplication of regulated gene Gene regulatory network in E. coli Model-2: Duplication of transcription factor Model-1 Model-2 32 TFs (out of 110) share regulated genes with a homologue 197 RG (out of 340) share transcription factors with a homologue 400 out of the 1233 (~1/3) interactions have homologous TFs sharing RG or homologous RGs sharing TFs. This suggests that duplication is a major mechanism for network growth 710 out of the 1233 interactions have either TF with a homologue or RG with a homologue but not share RG or TF, suggesting innovation of new edges or duplication followed by loss of regulatory interactions. 123 out of the 1233 interactions have no homologues for TFs and RG suggesting that these interactions were innovations.