Murcia_TP_Corrections
advertisement
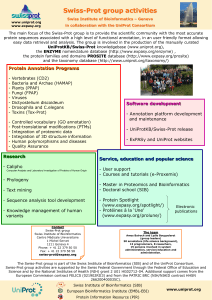
Corrections - The cacao genome is currently being sequenced http://www.ncbi.nlm.nih.gov/genomes/leuks.cgi - Human Chromosome 1 sequence Search ‘Genome’ with ‘homo sapiens chromosome 1’ RefSeq has an entry for each human chromosome (genome reference): these entries are numbered NC_000001 to NC_000024 Mapviewer view: http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?taxid=9606&chr=1 Entrez nucleotide view: http://www.ncbi.nlm.nih.gov/nuccore/224589800 - Measles virus complete genome (RefSeq) http://www.ncbi.nlm.nih.gov/nuccore/NC_001498 - Nucleic acid sequences available for human erythropoietin (EPO) (GenBank and RefSeq) http://www.ncbi.nlm.nih.gov/nucleotide/?term=homo+sapiens+erythropoietin+EPO mRNA coding for human EPO (in RefSeq) http://www.ncbi.nlm.nih.gov/nuccore/NM_000799.2 Look for Escherichia coli strain K-12 substrain W3110 complete genome (Query @ NCBI 'Genome') Query ‘Genome’ with ‘Escherichia coli strain K-12 substrain W3110’ http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genome&Cmd=ShowDetailView&TermToSearch=19221 CU466930.1 • http://www.ncbi.nlm.nih.gov/nuccore/167729007 Corresponding proteins in UniProtKB (query with CU466930): http://www.uniprot.org/uniprot/?query=CU466930&sort=score Corresponding proteins in NCBInr (query ‘nucleotide’ with CU466930 and follow the link to ‘protein’ http://www.ncbi.nlm.nih.gov/nuccore?Db=protein&DbFrom=nuccore&Cmd=Link&LinkNam e=nuccore_protein&IdsFromResult=167729007 AAFZ00000000.1 • http://www.ncbi.nlm.nih.gov/nuccore/60175893 No protein yet : no annotated CDS available (or not yet submitted by the authors)! EPO protein in different databases • UniProtKB/Swiss-Prot (reviewed) http://www.uniprot.org/uniprot/P01588 • UniProtKB/TrEMBL (unreviewed) http://www.uniprot.org/uniprot/B7ZKK5 • UniParc (follow the link from the UniProtKB sequence section) http://www.uniprot.org/uniparc/UPI0000033477 • NCBInr (see next slide) http://www.ncbi.nlm.nih.gov/protein/?term=homo+sapiens+erythropoietin+EPO • RefSeq (see next slide) http://www.ncbi.nlm.nih.gov/protein/NP_000790.2? • Ensembl (follow the link from the UniProt entry) http://www.ensembl.org/Homo_sapiens/Transcript/ProteinSummary?g=ENSG00000130427;r=7:100318423-100321323;t=ENST00000252723 http://www.uniprot.org/uniprot/P04150 - Which server? UniProt - Which database? UniProtKB/Swiss-Prot - What is the function of the protein? Receptor for glucocorticoids (GC). - How many different post-translational modifications (PTMs) ? Phosphorylated, Sumoylated, Ubiquitinated - How many phosphorylation sites? 10 sites - What are the associated GO terms? http://www.uniprot.org/uniprot/P04150#section_terms - What is the evidence for the existence of the protein(s)? http://www.uniprot.org/uniprot/P04150#section_attribute: at protein level - What is the corresponding mRNA? X03225 for example - How many different protein sequences are available for the corresponding gene? http://www.uniprot.org/uniprot/P04150#section_alternative: 9 isoforms http://www.ncbi.nlm.nih.gov/protein/NP_000167.1? - Which server? NCBI - Which database? RefSeq - What is the function of the protein? receptor for glucocorticoids - How many different post-translational modifications (PTMs) ? Phosphorylated, Sumoylated, Ubiquitinated - How many phosphorylation sites? 14 sites - What are the associated GO terms? Not directly available - What is the evidence for the existence of the protein(s)? This information is not available - What is the corresponding mRNA? ‘The reference sequence was derived from AC091925.3, X03225.1 and AC004782.1. ‘ - How many different protein sequences are available for the corresponding gene? This information is not directly available; go to Entrez Gene. http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=Retrieve&dopt=full_report&list_uids=2908 Find the UniProt entry corresponding to: - RefSeq NP_036231 - GI:584682 - Find the GI numbers corresponding to UniProtKB P04150 - Why are there so many GI numbers ? Find the UniProt entry corresponding to: - GI:584682 - Find the GI numbers corresponding to UniProtKB P04150 - Why are there so many GI numbers ? Find the GI numbers corresponding to UniProtKB P04150 Why are there so many GI numbers ? - Because there are several protein sequences corresponding to this gene due to alternative splicing events etc.; sequences have also been updated/modified; - The merging policy at NCBI (one sequence one entry) is not same as the one at UniProt (one entry one gene one species) Look for the URL corresponding to the following queries: - Mouse proteins localized in the nucleus Query: taxonomy:"Mus musculus [10090]" AND annotation:(type:location nucleus) URL:http://www.uniprot.org/uniprot/?query=taxonomy%3A%22Mus+musculus+[10090]%22+AND+annotation%3A%28type%3Alocation+nucleus%29&sort=score - Proteins for which a 3D structure is known (Hint: they have a cross-reference to PDB) Query: database:(type:pdb) URL: http://www.uniprot.org/uniprot/?query=database%3A%28type%3Apdb%29&sort=score - What is the query corresponding to this URL: http://www.uniprot.org/uniprot/?query=taxonomy%3A9606+AND+keyword%3A"complete+proteome“ Query: taxonomy:9606 AND keyword:"complete proteome“ -> human complete proteome - Modify the URL to get the same result, but for Escherichia coli (strain K12) Query: taxonomy:83333 AND keyword:"complete proteome“ URL: http://www.uniprot.org/uniprot/?query=taxonomy%3A83333+AND+keyword%3A"complete+proteome“ - Yeast (Saccharomyces cerevisiae) proteins found in the nucleus in UniProtKB/Swiss-Prot. Query: organism:4932 AND annotation:(type:location AND nucleus) AND reviewed:yes - How many of them have a nuclear localization which is 'experimentally proven' ? Query: organism:"Saccharomyces cerevisiae [4932]" AND annotation:(type:location nucleus confidence:experimental) AND reviewed:yes 1655 and 1392 entries respectively (query done in January, the 19th) - Download the list of corresponding accession numbers / protein names / gene names (use 'customize display'). - The set might not be complete: not all proteins have been tested to be localized in the nucleus ! And UniProtKB might not have annotated all the experiments showing that the yeast proteins are nuclear. P00001 @ UniProtKB P00001 @ NCBInr An 'old' publication cites a protein sequence with accession number (AC) o00597: Could you find it ? You did a proteomics analysis in December 2007 without any match. You repeat the analysis in April 2008 and get entry with AC P0C6S9 as the best match. Why ? P0C6S9 was first created in April 2008 in UniProtKB Compare the GO terms associated with mouse and human erythropoeitin (EPO) Have a look to the different GO evidence tags How many GO terms have been 'inferred by direct assay (IDA)' to the human EPO gene ? hierarchy of the GO term 'apoptosis' These GO terms are associated with insulin ! Several proteins have been identified in a proteomic experiment. Which GO terms do they share? (GI numbers of the identified proteins: 16130093, 20664033, 1789812, 89110178, 85677033, 27574045, 89111003, 229597766). GO terms in common… Searching databases with Blast The ‘alternative’ sequence(s) not ‘directly available’ for a lot of tools, including protein identification tools, Blast, depending on the server !…. Murcia, February, 2011 Protein Sequence Databases Blast P04150 against Swiss-Prot / homo sapiens @ UniProt Isoform sequences Murcia, February, 2011 Protein Sequence Databases Blast P04150 against Swiss-Prot / homo sapiens @ NCBI The isoform sequences (from Swiss-Prot) are not present in the NCBI protein database ! The .x number (P06401.4) correspond to the version number of the sequence…not to an alternatively spliced sequence ! Murcia, February, 2011 Protein Sequence Databases Blast A tool associated with the standard options to search sequences in UniProt databases The metal binding (Fe) site is conserved between HBB human and pea leghemoglobin! Detailed BLAST results Murcia, February, 2011 Protein Sequence Databases InterPro : other shema (Graphical view from UniProtKB) InterPro shema PFAM Graphical view Prosite Graphical view Not kepted in the InterPro overview ! Do a Blast with the sequence of the domain 'C-type lectin' of protein P28175 against UniProtKB/Swiss-Prot. Discuss the overview of your results. Look at the other domains present in the most similar sequences. Blast p28175 (complete seq) @ Swiss-Prot UniProt: Color code for identity scores (not alignment !) UniProt: Color code for identity scores (not alignment !) Blast p28175 (complete seq) @ NCBI against Swiss-Prot NCBI: Color key for alignment scores NCBI Swiss-Prot does not contain the alternative sequences (i.e. P28175-2) – !! NCBI gives the ‘version number’ of the Swiss-Prot sequence (i.e. Q8BU25.2)…. N-linked glycosylation (GlcNac): Look at the Swiss-Prot annotation (in a random ‘glycosylated’ entry) Query UniProtKB Taxonomic distribution TPNLINDTME Multiple alignment (ClustalW) -[LAPIQ]-N-[HAYRCS]-[ST]-[KLESGM] Logo N-glycosylation does not occur in Bacteria: …false positive ! 28 protein (within the set of 1000 proteins) are glycosylated according to the UniProtKB annotation…! Not easy to find that there is never a P after the N glycvosylation site (needs a lot of sequences….) ! C-x(2,4)-C-x(3)-[LIVMFYWC]-x(8)-H-x(3,5)-H Pattern scan The pattern missed a second Zn finger in the same protein i.e. Q24174 Pattern Profile The pattern: C - X(2,4) - C - X(3) - [LIVMFYWC] - X(8) - H - X(3,5) – H Should includes: YRCVLCGTVAKSRNSLHSHMSrQHRGIST C-X(2,4)-C-X(3)-[LIVMFYWCA]-X(8)-H-X(3,5)-H Yes ! But: The pattern becomes less restrictive. But you get more sequences which should not be here. As the results are limited to 1000, the number of hits is not the same… Discriminators (Signatures, descriptors) for the Zinc finger C2H2 type domain can be found in Prosite (Pattern and Profile) and Pfam (HMM) Doublecortin Kinase The doublecortin domain is associated with many different domains (not only kinase) Seq 1 Seq 2 Patient with cardiovascular disease Lost of the protein kinase ATP binding site ! Step 1: scan UniProtKB/Swiss-Prot with the pattern Use the ‘scanprosite’ tool at http://www.expasy.org/tools/scanprosite/ At the bottom of the Scan prosite result page: Step 2: Retrieve the 103 human entries @ UniProt (go at the bottom of the Scan Prosite result page; Matched UniProtKB entries) Step 3: Retrieve the sequences annotated as being ‘phosphorylated on a Thr’ -> 19 candidates to be manually checked …. The end