Protein Synthesis
advertisement

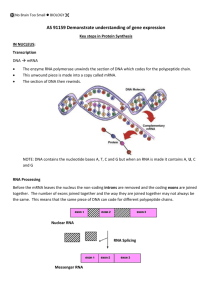
The Central Dogma of Life. replication Protein Synthesis • The information content of DNA is in the form of specific sequences of nucleotides along the DNA strands • The DNA inherited by an organism leads to specific traits by dictating the synthesis of proteins • The process by which DNA directs protein synthesis, gene expression includes two stages, called transcription and translation Transcription and Translation • Cells are governed by a cellular chain of command – DNA RNA protein • Transcription – Is the synthesis of RNA under the direction of DNA – Produces messenger RNA (mRNA) • Translation – Is the actual synthesis of a polypeptide, which occurs under the direction of mRNA – Occurs on ribosomes Transcription and Translation • In prokaryotes transcription and translation occur together TRANSCRIPTION DNA mRNA Ribosome TRANSLATION Polypeptide Figure 17.3a (a) Prokaryotic cell. In a cell lacking a nucleus, mRNA produced by transcription is immediately translated without additional processing. Transcription and Translation • In a eukaryotic cell the nuclear envelope separates transcription from translation • Extensive RNA processing occurs in the nucleus Nuclear envelope DNA TRANSCRIPTION Pre-mRNA RNA PROCESSING mRNA Ribosome TRANSLATION Polypeptide (b) Eukaryotic cell. The nucleus provides a separate compartment for transcription. The original RNA transcript, called pre-mRNA, is processed in various ways before leaving the nucleus as mRNA. Transcription • Transcription is the DNAdirected synthesis of RNA • RNA synthesis – Is catalyzed by RNA polymerase, which pries the DNA strands apart and hooks together the RNA nucleotides – Follows the same basepairing rules as DNA, except that in RNA, uracil substitutes for thymine RNA • RNA is single stranded, not double stranded like DNA • RNA is short, only 1 gene long, where DNA is very long and contains many genes • RNA uses the sugar ribose instead of deoxyribose in DNA • RNA uses the base uracil (U) instead of thymine (T) in DNA. Table 17.1 Synthesis of an RNA Transcript • The stages of transcription are – Initiation – Elongation – Termination Promoter Transcription unit 5 3 3 5 Start point RNA polymerase DNA Initiation. After RNA polymerase binds to the promoter, the DNA strands unwind, and the polymerase initiates RNA synthesis at the start point on the template strand. 1 5 3 Unwound DNA 3 5 Template strand of DNA transcript 2 Elongation. The polymerase moves downstream, unwinding the DNA and elongating the RNA transcript 5 3 . In the wake of transcription, the DNA strands re-form a double helix. Rewound RNA RNA 5 3 3 5 3 5 RNA transcript 3 Termination. Eventually, the RNA transcript is released, and the polymerase detaches from the DNA. 5 3 3 5 5 Completed RNA transcript 3 Synthesis of an RNA Transcript - Initiation • Promoters signal the initiation of RNA synthesis • Transcription factors help eukaryotic RNA polymerase recognize promoter sequences 1 Eukaryotic promoters TRANSCRIPTION DNA RNA PROCESSING Pre-mRNA mRNA TRANSLATION Ribosome Polypeptide Promoter 5 3 3 5 T A T A A AA AT AT T T T TATA box Start point Template DNA strand Several transcription factors 2 Transcription factors 5 3 3 5 3 Additional transcription • A crucial promoter DNA sequence is called a TATA box. factors RNA polymerase II 5 3 Transcription factors 3 5 5 RNA transcript Transcription initiation complex Synthesis of an RNA Transcript - Elongation • • RNA polymerase synthesizes a single strand of RNA against the DNA template strand (anti-sense strand), adding nucleotides to the 3’ end of the RNA chain As RNA polymerase moves along the DNA it continues to untwist the double helix, exposing about 10 to 20 DNA bases at a time for pairing with RNA nucleotides Non-template strand of DNA Elongation RNA nucleotides RNA polymerase A 3 T C C A A 3 end U 5 A E G C A T A G G T T Direction of transcription (“downstream”) 5 Newly made RNA Template strand of DNA Synthesis of an RNA Transcript - Termination • • Specific sequences in the DNA signal termination of transcription When one of these is encountered by the polymerase, the RNA transcript is released from the DNA and the double helix can zip up again. Transcription Overview Post Termination RNA Processing • Most eukaryotic mRNAs aren’t ready to be translated into protein directly after being transcribed from DNA. mRNA requires processing. • Transcription of RNA processing occur in the nucleus. After this, the messenger RNA moves to the cytoplasm for translation. • The cell adds a protective cap to one end, and a tail of A’s to the other end. These both function to protect the RNA from enzymes that would degrade • Most of the genome consists of non-coding regions called introns • – Non-coding regions may have specific chromosomal functions or have regulatory purposes – Introns also allow for alternative RNA splicing Thus, an RNA copy of a gene is converted into messenger RNA by doing 2 things: – Add protective bases to the ends – Cut out the introns Alteration of mRNA Ends • Each end of a pre-mRNA molecule is modified in a particular way – The 5 end receives a modified nucleotide cap – The 3 end gets a poly-A tail A modified guanine nucleotide added to the 5 end TRANSCRIPTION RNA PROCESSING 50 to 250 adenine nucleotides added to the 3 end DNA Pre-mRNA 5 mRNA Protein-coding segment Polyadenylation signal 3 G P P P AAUAAA AAA…AAA Ribosome TRANSLATION 5 Cap Polypeptide 5 UTR Start codon Stop codon 3 UTR Poly-A tail RNA Processing - Splicing • The original transcript from the DNA is called pre-mRNA. • It contains transcripts of both introns and exons. • The introns are removed by a process called splicing to produce messenger RNA (mRNA) RNA Processing - Splicing • Ribozymes are catalytic RNA molecules that function as enzymes and can splice RNA • RNA splicing removes introns and joins exons 5 Exon Intron TRANSCRIPTION DNA Exon 3 Pre-mRNA 5 Cap 1 RNA PROCESSING Intron Exon Poly-A tail 30 31 104 105 146 Pre-mRNA Coding segment mRNA Ribosome Introns cut out and exons spliced together TRANSLATION Polypeptide mRNA 5 Cap 1 3 UTR Figure 17.10 Poly-A tail 146 3 UTR RNA Processing • RNA Splicing can also be carried out by spliceosomes RNA transcript (pre-mRNA) 5 Intron Exon 1 Exon 2 Protein 1 Other proteins snRNA snRNPs Spliceosome 2 5 Spliceosome components 3 5 mRNA Exon 1 Exon 2 Cut-out intron Alternative Splicing (of Exons) • How is it possible that there are millions of human antibodies when there are only about 30,000 genes? • Alternative splicing refers to the different ways the exons of a gene may be combined, producing different forms of proteins within the same gene-coding region • Alternative pre-mRNA splicing is an important mechanism for regulating gene expression in higher eukaryotes RNA Processing • Proteins often have a modular architecture consisting of discrete structural and functional regions called domains • In many cases different exons code for the different domains in a protein Gene DNA Exon 1 Exon 2 Intron Intron Exon 3 Transcription RNA processing Translation Domain 3 Domain 2 Domain 1 Figure 17.12 Polypeptide Translation • Translation is the RNAdirected synthesis of a polypeptide • Translation involves – – – – TRANSCRIPTION DNA mRNA Ribosome TRANSLATION Polypeptide Amino acids Polypeptide mRNA Ribosomes - Ribosomal RNA Transfer RNA Genetic coding - codons tRNA with amino acid Ribosome attached Gly tRNA Anticodon A A A U G G U U U G G C Codons 5 mRNA 3 The Genetic Code • Genetic information is encoded as a sequence of nonoverlapping base triplets, or codons • The gene determines the sequence of bases along the length of an mRNA molecule Gene 2 DNA molecule Gene 1 Gene 3 DNA strand (template) 3 A C C A A A C C G A G T 5 TRANSCRIPTION mRNA 5 U G G U U U G G C U C A Codon TRANSLATION Protein Trp Amino acid Phe Gly Ser 3 The Genetic Code • Codons: 3 base code for the production of a specific amino acid, sequence of three of the four different nucleotides • Since there are 4 bases and 3 positions in each codon, there are 4 x 4 x 4 = 64 possible codons • 64 codons but only 20 amino acids, therefore most have more than 1 codon • 3 of the 64 codons are used as STOP signals; they are found at the end of every gene and mark the end of the protein • One codon is used as a START signal: it is at the start of every protein • Universal: in all living organisms The Genetic Code Second mRNA base U C A UAU UUU UCU Tyr Phe UAC UUC UCC U UUA UCA Ser UAA Stop UAG Stop UUG Leu UCG CUU CUC C CUA CUG CCU CCC Leu CCA CCG Pro AUU AUC A AUA AUG ACU ACC ACA ACG Thr GUU G GUC GUA GUG lle Met or start GCU GCC Val GCA GCG Ala G U UGU Cys UGC C UGA Stop A UGG Trp G U CAU CGU His CAC CGC C Arg CAA CGA A Gln CAG CGG G U AAU AGU Asn AAC AGC Ser C A AAA AGA Lys Arg G AAG AGG U GAU GGU C GAC Asp GGC Gly GAA GGA A Glu GAG GGG G Third mRNA base (3 end) First mRNA base (5 end) • A codon in messenger RNA is either translated into an amino acid or serves as a translational start/stop signal Transfer RNA • Consists of a single RNA strand that is only about 80 nucleotides long • Each carries a specific amino acid on one end and has an anticodon on the other end • A special group of enzymes pairs up the proper tRNA molecules with their corresponding amino acids. • tRNA brings the amino acids to the ribosomes, 3 A C C A 5 C G The “anticodon” is the 3 RNA bases that G C C G matches the 3 bases of the codon on the U G U A mRNA molecule A U A U U C UA C A C AG * G * U G U G C C * * * U C * * G AG C (a) Two-dimensional structure. The four base-paired regions and three G C U A loops are characteristic of all tRNAs, as is the base sequence of the * G amino acid attachment site at the 3 end. The anticodon triplet is A A* C unique to each tRNA type. (The asterisks mark bases that have been U * chemically modified, a characteristic of tRNA.) A G A Amino acid attachment site Anticodon C U C G A G A G * * G A G G Hydrogen bonds Transfer RNA • 3 dimensional tRNA molecule is roughly “L” shaped 5 3 Amino acid attachment site Hydrogen bonds A AG 3 Anticodon (b) Three-dimensional structure Anticodon 5 (c) Symbol used in the book Ribosomes • Ribosomes facilitate the specific coupling of tRNA anticodons with mRNA codons during protein synthesis • The 2 ribosomal subunits are constructed of proteins and RNA molecules named ribosomal RNA or rRNA DNA TRANSCRIPTION mRNA Ribosome TRANSLATION Polypeptide Growing polypeptide Exit tunnel tRNA molecules Large subunit E P A Small subunit 5 mRNA 3 (a) Computer model of functioning ribosome. This is a model of a bacterial ribosome, showing its overall shape. The eukaryotic ribosome is roughly similar. A ribosomal subunit is an aggregate of ribosomal RNA molecules and proteins. Ribosome • The ribosome has three binding sites for tRNA – The P site – The A site – The E site P site (Peptidyl-tRNA binding site) A site (AminoacyltRNA binding site) E site (Exit site) Large subunit E mRNA binding site P A Small subunit (b) Schematic model showing binding sites. A ribosome has an mRNA binding site and three tRNA binding sites, known as the A, P, and E sites. This schematic ribosome will appear in later diagrams. Building a Polypeptide Amino end Growing polypeptide Next amino acid to be added to polypeptide chain tRNA 3 mRNA 5 Codons (c) Schematic model with mRNA and tRNA. A tRNA fits into a binding site when its anticodon basepairs with an mRNA codon. The P site holds the tRNA attached to the growing polypeptide. The A site holds the tRNA carrying the next amino acid to be added to the polypeptide chain. Discharged tRNA leaves via the E site. Building a Molecule of tRNA • A specific enzyme called an aminoacyl-tRNA synthetase joins each amino acid to the correct tRNA Amino acid P P Aminoacyl-tRNA synthetase (enzyme) 1 Active site binds the amino acid and ATP. P Adenosine ATP 2 ATP loses two P groups and joins amino acid as AMP. P Pyrophosphate Pi Phosphates P Adenosine Pi Pi tRNA 3 Appropriate tRNA covalently Bonds to amino Acid, displacing AMP. P Adenosine AMP 4 Activated amino acid is released by the enzyme. Figure 17.15 Aminoacyl tRNA (an “activated amino acid”) Building a Polypeptide • We can divide translation into three stages – Initiation – Elongation – Termination • The AUG start codon is recognized by methionyl-tRNA or Met • Once the start codon has been identified, the ribosome incorporates amino acids into a polypeptide chain • RNA is decoded by tRNA (transfer RNA) molecules, which each transport specific amino acids to the growing chain • Translation ends when a stop codon (UAA, UAG, UGA) is reached Initiation of Translation • The initiation stage of translation brings together mRNA, tRNA bearing the first amino acid of the polypeptide, and two subunits of a ribosome Large ribosomal subunit P site 3 U A C 5 5 A U G 3 Initiator tRNA GTP GDP E A mRNA 5 Start codon mRNA binding site 1 5 3 Small ribosomal subunit A small ribosomal subunit binds to a molecule of mRNA. In a prokaryotic cell, the mRNA binding site on this subunit recognizes a specific nucleotide sequence on the mRNA just upstream of the start codon. An initiator tRNA, with the anticodon UAC, base-pairs with the start codon, AUG. This tRNA carries the amino acid methionine (Met). 3 Translation initiation complex 2 The arrival of a large ribosomal subunit completes the initiation complex. Proteins called initiation factors (not shown) are required to bring all the translation components together. GTP provides the energy for the assembly. The initiator tRNA is in the P site; the A site is available to the tRNA bearing the next amino acid. Elongation of the Polypeptide Chain • In the elongation stage, amino acids are added one by one to the preceding amino acid TRANSCRIPTION 1 Codon recognition. The anticodon of an incoming aminoacyl tRNA base-pairs with the complementary mRNA codon in the A site. Hydrolysis of GTP increases the accuracy and efficiency of this step. Amino end of polypeptide DNA mRNA Ribosome TRANSLATION Polypeptide E mRNA Ribosome ready for next aminoacyl tRNA 5 3 P A site site 2 GTP 2 GDP E E P P A GDP 3 Translocation. The ribosome translocates the tRNA in the A site to the P site. The empty tRNA in the P site is moved to the E site, where it is released. The mRNA moves along with its bound tRNAs, bringing the next codon to be translated into the A site. GTP E P A A 2 Peptide bond formation. An rRNA molecule of the large subunit catalyzes the formation of a peptide bond between the new amino acid in the A site and the carboxyl end of the growing polypeptide in the P site. This step attaches the polypeptide to the tRNA in the A site. Termination of Translation • The final step in translation is termination. When the ribosome reaches a STOP codon, there is no corresponding transfer RNA. Instead, a small protein called a “release factor” attaches to the stop codon. The release factor causes the whole complex to fall apart: messenger RNA, the two ribosome subunits, the new polypeptide. The messenger RNA can be translated many times, to produce many protein copies. • • • Release factor Free polypeptide 5 3 3 5 5 3 Stop codon (UAG, UAA, or UGA) 1 When a ribosome reaches a stop 2 The release factor hydrolyzes 3 The two ribosomal subunits codon on mRNA, the A site of the the bond between the tRNA in and the other components of ribosome accepts a protein called the P site and the last amino the assembly dissociate. a release factor instead of tRNA. acid of the polypeptide chain. The polypeptide is thus freed from the ribosome. Translation: Initiation • mRNA binds to a ribosome, and the transfer RNA corresponding to the START codon binds to this complex. Ribosomes are composed of 2 subunits (large and small), which come together when the messenger RNA attaches during the initiation process. Translation: Elongation • Elongation: the ribosome moves down the messenger RNA, adding new amino acids to the growing polypeptide chain. • The ribosome has 2 sites for binding transfer RNA. The first RNA with its attached amino acid binds to the first site, and then the transfer RNA corresponding to the second codon bind to the second site. Translation: Elongation • The ribosome then removes the amino acid from the first transfer RNA and attaches it to the second amino acid. • At this point, the first transfer RNA is empty: no attached amino acid, and the second transfer RNA has a chain of 2 amino acids attached to it. Translation: Termination • The elongation cycle repeats as the ribosome moves down the messenger RNA, translating it one codon and one amino acid at a time. • The process repeats until a STOP codon is reached. Polyribosomes • A number of ribosomes can translate a single mRNA molecule simultaneously forming a polyribosome • Polyribosomes enable a cell to make many copies of a polypeptide very quickly Completed polypeptide Growing polypeptides Incoming ribosomal subunits Start of End of mRNA mRNA (5 end) (3 end) (a) An mRNA molecule is generally translated simultaneously by several ribosomes in clusters called polyribosomes. Ribosomes mRNA 0.1 µm This micrograph shows a large polyribosome in a prokaryotic cell (TEM). Comparing Gene Expression In Prokaryotes And Eukaryotes • In a eukaryotic cell: – – • The nuclear envelope separates transcription from translation Extensive RNA processing occurs in the nucleus Prokaryotic cells lack a nuclear envelope, allowing translation to begin while transcription progresses RNA polymerase DNA mRNA Polyribosome RNA polymerase Direction of transcription 0.25 mm DNA Polyribosome Polypeptide (amino end) Ribosome mRNA (5 end) A summary of transcription and translation in a eukaryotic cell DNA TRANSCRIPTION 1RNA is transcribed from a DNA template. 3 RNA transcript 5 RNA polymerase Exon RNA PROCESSING 2 In eukaryotes, the RNA transcript (premRNA) is spliced and modified to produce mRNA, which moves from the nucleus to the cytoplasm. RNA transcript (pre-mRNA) Intron Aminoacyl-tRNA synthetase NUCLEUS Amino acid FORMATION OF INITIATION COMPLEX CYTOPLASM AMINO ACID ACTIVATION tRNA 3 After leaving the 4 Each amino acid attaches to its proper tRNA with the help of a specific enzyme and ATP. nucleus, mRNA attaches to the ribosome. mRNA Growing polypeptide Activated amino acid Ribosomal subunits 5 TRANSLATION 5 E A A A A U G G U U U A U G Figure 17.26 Codon Ribosome Anticodon A succession of tRNAs add their amino acids to the polypeptide chain as the mRNA is moved through the ribosome one codon at a time. (When completed, the polypeptide is released from the ribosome.) Post-translation • The new polypeptide is now floating loose in the cytoplasm if translated by a free ribosome. • Polypeptides fold spontaneously into their active configuration, and they spontaneously join with other polypeptides to form the final proteins. • Often translation is not sufficient to make a functional protein, polypeptide chains are modified after translation • Sometimes other molecules are also attached to the polypeptides: sugars, lipids, phosphates, etc. All of these have special purposes for protein function. Targeting Polypeptides to Specific Locations • Completed proteins are targeted to specific sites in the cell • Two populations of ribosomes are evident in cells: free ribsomes (in the cytosol) and bound ribosomes (attached to the ER) – – Free ribosomes mostly synthesize proteins that function in the cytosol Bound ribosomes make proteins of the endomembrane system and proteins that are secreted from the cell • Ribosomes are identical and can switch from free to bound Targeting Polypeptides to Specific Locations • • • • • Polypeptide synthesis always begins in the cytosol Synthesis finishes in the cytosol unless the polypeptide signals the ribosome to attach to the ER Polypeptides destined for the ER or for secretion are marked by a signal peptide A signal-recognition particle (SRP) binds to the signal peptide The SRP brings the signal peptide and its ribosome to the ER Ribosomes mRNA Signal peptide Signalrecognition particle SRP receptor (SRP) protein CYTOSOL ER LUMEN Translocation complex ER membrane Signal peptide removed Protein Mutation Causes and Rate • The natural replication of DNA produces occasional errors. DNA polymerase has an editing mechanism that decreases the rate, but it still exists • Typically genes incur base substitutions about once in every 10,000 to 1,000,000 cells • Since we have about 6 billion bases of DNA in each cell, virtually every cell in your body contains several mutations • Mutations can be harmful, lethal, helpful, silent • However, most mutations are neutral: have no effect • Only mutations in cells that become sperm or eggs— are passed on to future generations • Mutations in other body cells only cause trouble when they cause cancer or related diseases Mutagens • Mutagens are chemical or physical agents that interact with DNA to cause mutations. • Physical agents include high-energy radiation like X-rays and ultraviolet light • Chemical mutagens fall into several categories. – – – Chemicals that are base analogues that may be substituted into DNA, but they pair incorrectly during DNA replication. Interference with DNA replication by inserting into DNA and distorting the double helix. Chemical changes in bases that change their pairing properties. • Tests are often used as a preliminary screen of chemicals to identify those that may cause cancer • Most carcinogens are mutagenic and most mutagens are carcinogenic. Viral Mutagens • Scientists have recognized a number of tumor viruses that cause cancer in various animals, including humans • About 15% of human cancers are caused by viral infections that disrupt normal control of cell division • All tumor viruses transform cells into cancer cells through the integration of viral nucleic acid into host cell DNA. Point mutations • Point mutations involve alterations in the structure or location of a single gene. Generally, only one or a few base pairs are involved. • Point mutations can signficantly affect protein structure and function • Point mutations may be caused by physical damage to the DNA from radiation or chemicals, or may occur spontaneously • Point mutations are often caused by mutagens Point Mutation • The change of a single nucleotide in the DNA’s template strand leads to the production of an abnormal protein Wild-type hemoglobin DNA 3 Mutant hemoglobin DNA 5 C T T In the DNA, the mutant template strand has an A where the wild-type template has a T. G U A The mutant mRNA has a U instead of an A in one codon. 3 5 T C A mRNA mRNA G A A 5 3 5 3 Normal hemoglobin Sickle-cell hemoglobin Glu Val The mutant (sickle-cell) hemoglobin has a valine (Val) instead of a glutamic acid (Glu). Types of Point Mutations • Point mutations within a gene can be divided into two general categories – Base-pair substitutions - is the replacement of one nucleotide and its partner with another pair of nucleotides – Base-pair insertions or deletions - are additions or losses of nucleotide pairs in a gene Base-Pair Substitutions • • • Silent - changes a codon but codes for the same amino acid Missense - substitutions that change a codon for one amino acid into a codon for a different amino acid Nonsense -substitutions that change a codon for one amino acid into a stop codon Wild type mRNA Protein A U G 5 Met A A G U U U G G C U A A Lys Phe Gly 3 Stop Amino end Carboxyl end Base-pair substitution No effect on amino acid sequence U instead of C A U G A A G U U U G G U U A A Met Lys Missense Phe Gly Stop A instead of G A U G A A G U U U A G U U A A Met Lys Phe Ser Stop Nonsense U instead of A A U G U A G U U U G G C U A A Met Stop Insertions and Deletions – Are additions or losses of nucleotide pairs in a gene – May produce frameshift mutations that will change the reading frame of the gene, and alter all codons downstream from the mutation. Wild type mRNA Protein 5 A U GA A GU U U G G C U A A Met Lys Gly Phe Stop Amino end Carboxyl end Base-pair insertion or deletion Frameshift causing immediate nonsense Extra U AU G U A AG U U U G GC U A Met Stop Frameshift causing extensive missense U Missing A U G A A GU U G G C U A A Met Lys Leu Ala Insertion or deletion of 3 nucleotides: no frameshift but extra or missing amino acid A A G Missing A U G U U U G G C U A A Met Phe Gly Stop 3