Chapter 24: Bacteria & Viruses
advertisement

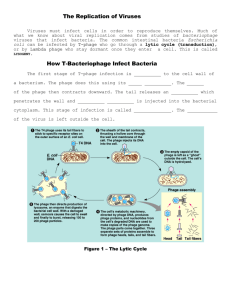
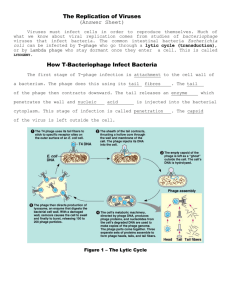
Viruses and Prokaryotes Chapter 24 Learning Objective 1 • What is the structure of a virus? • Contrast a virus with a living cell Virus (Virion) • Subcellular particle • Consists of • • DNA or RNA genome surrounded by protein coat (capsid) Virus Structure RNA inside capsid Capsid 0.1 µm Fig. 24-1a, p. 502 Capsid with antenna-like fibers DNA inside capsid 0.05 µm Fig. 24-1b, p. 502 DNA inside capsid Capsid Tail Tail fibers Emerging DNA 0.1 µm Fig. 24-1c, p. 502 Viruses • Cannot metabolize on their own • Contain nucleic acids necessary to make copies of themselves • but must invade and use metabolic machinery of living cells in order to reproduce KEY CONCEPTS • A virus is a small particle consisting of a DNA or RNA genome surrounded by a protein coat Learn more about virus structure by clicking on the figure in ThomsonNOW. Learning Objective 2 • According to current hypotheses, what is the evolutionary origin of viruses? Origin of Viruses • Viruses may be bits of nucleic acid that originally “escaped” from animal, plant, or bacterial cells Hypothesis • Viruses must have evolved before the three domains diverged • It is unlikely that similar viruses that infect archaea and bacteria evolved twice Learning Objective 3 • Characterize bacteriophages (phages) • • viruses that infect bacteria What is the difference between a lytic cycle and a lysogenic cycle? Viral Reproductive Cycles • Lytic cycle • virus destroys host cell • Temperate viruses • do not always destroy their hosts • Lysogenic cycle • viral genome replicated along with host DNA Lytic Cycle • 5 steps: • • • • • attachment to host cell penetration of viral nucleic acid into host cell replication of viral nucleic acid assembly of components into new viruses release from host cell Lytic Cycle Phages Bacterium 1 Attachment. Phage attaches to cell surface of bacterium. Bacterial DNA 2 Penetration. Phage DNA enters bacterial cell. Phage protein Phage DNA 3 Replication and synthesis. Phage DNA is replicated. Phage proteins are synthesized. Fig. 24-2a (1), p. 504 4 Assembly. Phage components are assembled into new viruses. 5 Release. Bacterial cell lyses and releases many phages that can then infect other cells. Fig. 24-2a (2), p. 504 0.25 µm Fig. 24-2b, p. 504 Lysogenic Cycle • Prophage • • Lysogenic cells • • nucleic acid of phage becomes integrated into bacterial DNA bacterial cells that carry prophages Lysogenic conversion • bacterial cells containing certain temperate viruses exhibit new properties Lysogenic Cycle 1 Attachment. Phage attaches to cell surface of bacterium. 2 Penetration. Phage DNA enters bacterial cell. Prophage 3 Integration. Phage DNA integrates into bacterial DNA. These cells may exhibit new properties. 4 Replication. Integrated prophage replicates when bacterial DNA replicates. Fig. 24-3, p. 504 KEY CONCEPTS • Evolution occurs rapidly in prokaryotes; natural selection acts on the genetic variation provided by mutations and genetic recombination and is facilitated by rapid reproduction Insert “The two different ways that viruses replicate (lytic and lysogenic cycles)” Tbd *suggested by Mary Durant, who will review existing animations currently slated for pickup (cd) Watch the lytic and lysogenic cycles by clicking on the figure in ThomsonNOW. Learning Objective 4 • Compare viral infection of animals and plants • Identify specific diseases caused by animal viruses Animal Viruses • Viruses enter animal cells by membrane fusion or endocytosis • Viral nucleic acid replicated in host cell • • proteins synthesized new viruses assembled and released from cell Envelope proteins 1 Virus attaches to specific receptors on plasma membrane of host cell. Envelope Capsid Nucleic acid Membrane Fusion 2 Membrane fusion. Viral envelope fuses with plasma membrane. Receptors Host-cell plasma membrane Cytoplasm Capsid Nucleus Nucleic acid Ribosomes ER mRNA 10 Viruses are released from host cell. 3 Virus is released into host-cell cytoplasm. 4 Viral nucleic acid separates from its capsid. 5 Viral nucleic acid enters host-cell nucleus and replicates. 6 Viral nucleic acid is transcribed into mRNA. 7 Host ribosomes are directed by mRNA to synthesize viral proteins. 8 Vesicles transport glycoproteins to hostcell plasma membrane. New viruses are assembled and enveloped 9 by host-cell plasma membrane. Fig. 24-4b, p. 508 Endocytosis 3 Endosomal vesicle forms and moves into cytoplasm. 2 Host-cell plasma membrane surrounds virus. 1 Virus makes contact with plasma membrane of host cell. Host-cell cytoplasm Host-cell plasma membrane 4 Virus is released into host-cell cytoplasm. 5 Viral envelope fuses with host-cell plasma membrane (not shown). Fig. 24-4c, p. 508 Viral Diseases • DNA viruses cause • • smallpox, herpes, respiratory infections, gastrointestinal disorders RNA viruses cause • influenza, upper respiratory infections, AIDS, some types of cancer Rubella • An RNA virus Plant Viruses • Mostly RNA viruses • Spread among plants by insect vectors • Spread through plant via plasmodesmata Plant Viruses Learning Objective 5 • Describe the reproductive cycle of a retrovirus, such as human immunodeficiency virus (HIV) Retroviruses • Use reverse transcriptase • Transcribe RNA genome into DNA intermediate • • becomes integrated into host DNA Synthesize copies of viral RNA HIV Envelope protein Envelope Capsid Enzymes (reverse transcriptase, ribonuclease, integrase, protease) Host-cell plasma membrane HIV Nucleic acid (RNA) 1 HIV attaches to host-cell plasma membrane. 2 HIV enters host-cell cytoplasm. CD4 Receptors Viral nucleic Reverse acid (RNA) transcriptase Cytoplasm ssDNA Nucleus Host chromosome Viral RNA dsDNA 3 Capsid is removed by enzymes. Reverse transcriptase catalyzes synthesis of single-stranded (ss) DNA that is complementary to viral RNA. 4 The DNA strand then serves as template for synthesis of complementary DNA strand, resulting in double-stranded (ds) DNA. 5 dsDNA is transferred to host nucleus and enzyme integrase integrates DNA into host chromosome. 6 When activated, viral DNA uses host enzymes to transcribe viral RNA. 7 Viral RNA leaves nucleus, viral proteins are synthesized on host ribosomes, and virus is assembled. 8 Virus buds from host cell, using host-cell plasma membrane to make viral envelope. Fig. 24-5, p. 509 Watch the HIV life cycle by clicking on the figure in ThomsonNOW Learning Objective 6 • What are viroids and prions? Viroids and Prions • Viroids • • short strands of RNA with no protein coat Prions • • consists only of protein cause transmissible spongiform encephalopathies (TSEs) Prions Contacts Prion Normal protein (PrP) 1 Prion induces normal PrP to misfold, forming another prion. Contacts Contacts 2 Each prion can induce additional PrP proteins to misfold. 3 Proteins aggregate. Fig. 24-7, p. 511 Learning Objective 7 • Describe the structure and common shapes of prokaryotic cells Prokaryotic Cells • Do not have membrane-enclosed organelles • such as nuclei and mitochondria Prokaryotic Cell Outer membrane Pili (structures used for attachment) Peptidoglycan layer Cell wall Nuclear area (nucleoid) Storage granule Plasmid (DNA) Flagellum Ribosomes Bacterial chromosome (DNA) Capsule Plasma membrane Fig. 24-9, p. 513 Bacterial Shapes • Spherical (cocci) Bacterial Shapes • Rod shaped (bacilli) Bacterial Shapes • Spiral • • rigid helix (spirillum) flexible helix (spirochete) Bacteria Structure • Cell walls composed of peptidoglycan • Some have capsule surrounding cell wall Bacterial Cell Walls • Gram-positive bacteria • • • walls very thick consist mainly of peptidoglycan Gram-negative bacteria • • walls have thin peptidoglycan layer outer membrane (like plasma membrane) Gram-Positive Cell Wall Gram-Negative Cell Wall Thick peptidoglycan layer Cell wall Plasma membrane (inner membrane) Transport protein (a) Gram-positive cell wall. Fig. 24-10a, p. 514 Polysaccharides Lipoprotein Outer membrane Cell wall Thin peptidoglycan layer Plasma membrane Transport protein (b) Gram-negative cell wall. Fig. 24-10b, p. 514 Bacterial Pili • Protein structures extending from cell • help bacteria adhere to one another or to other surfaces Bacterial Flagella • Different from eukaryotic flagella • Consist of • • • • basal body hook filament Produce rotary motion Bacterial Flagella Plasma membrane Cytoplasm Basal body Peptidoglycan layer Outer membrane Protein rings Hook Filament Fig. 24-11b, p. 515 Learn more about the structure of prokaryotes and their cell walls by clicking on the figures in ThomsonNOW Learning Objective 8 • Describe asexual reproduction in prokaryotes • Summarize three mechanisms (transformation, transduction, and conjugation) that may lead to genetic recombination Prokaryote Genes • Genetic material consists of • • 1 circular DNA molecule 1 or more plasmids (circular DNA fragments) Asexual Reproduction • Binary fission • • Budding • • cell divides, forming two cells bud forms, separates from mother cell Fragmentation • • walls form inside cell separates into several cells Genetic Material Exchange • Transformation • • Transduction • • bacterial cell takes in DNA fragments released by another cell phage carries bacterial DNA from one bacterial cell into another Conjugation • two cells of different mating types exchange genetic material Transformation 1 Bacterium dies and releases DNA. 2 Fragments of foreign DNA bind to proteins on surface of living bacterium. 3 DNA enters cell, and some DNA is incorporated into host cell by reciprocal recombination. DNA exchanged Fig. 24-12, p. 515 Transduction 1 DNA of a phage penetrates bacterial cell. 2 Phage DNA may become integrated with host-cell DNA as a prophage. Phage DNA with bacterial genes Fragmented bacterial DNA 3 When the prophage becomes lytic, bacterial DNA is degraded and new phages are produced. New phages may contain some bacterial DNA. Fig. 24-13a, p. 516 4 Bacterial cell lyses and releases many phages, which can then infect other cells. 5 Phage infects new host cell. 6 Bacterial genes introduced into new host cell are integrated into host's DNA. They become a part of bacterial DNA and are replicated along with it. Fig. 24-13b, p. 516 1 DNA of a phage penetrates bacterial cell. 2 Phage DNA may become integrated with hostcell DNA as a prophage. 3 When the prophage becomes lytic, bacterial DNA is degraded and new phages are produced. New phages may contain some bacterial DNA. 4 Bacterial cell lyses and releases many phages, which can then infect other cells. 5 Phage infects new host cell. 6 Bacterial genes introduced into new host cell are integrated into host's DNA. They become a part of bacterial DNA and are replicated along with it. Stepped Art Phage DNA with bacterial genes Fragmented bacterial DNA Fig. 24-13b, p. 516 Conjugation F+ (donor) cell F– (recipient) cell 1 F+ (donor) cell produces sex pilus. Bacterial chromosome F plasmid 2 Sex pilus develops into conjugation bridge. 3 DNA replicates, and single strand of F plasmid DNA is transferred from F+ cell to F– cell. Both bacterial cells now 4 contain double-stranded F plasmid. The F– cell has been converted to an F+ cell. Fig. 24-14b, p. 517 . Learning Objective 9 • What are the modes of nutrition and metabolic adaptations of prokaryotes? Prokaryote Nutrition • Most are heterotrophs • • obtain energy and carbon from other organisms Some are autotrophs • make their own organic molecules from simple raw materials Heterotrophs • Chemoheterotrophs • • • free-living decomposers obtain carbon and energy from dead organic matter Photoheterotrophs • • obtain carbon from other organisms photosynthetic pigments trap light energy Autotrophs • Photoautotrophs • • obtain energy from sunlight Chemoautotrophs • obtain energy by oxidizing inorganic chemicals such as ammonia Aerobes and Anaerobes • Aerobic bacteria • • Facultative anaerobes • • require oxygen for cellular respiration metabolize anaerobically when necessary Obligate anaerobes • only metabolize anaerobically Learning Objective 10 • Compare the three domains: Archaea, Bacteria, and Eukarya 3 Domains • Domain Archaea (prokaryotes) • • Domain Bacteria (prokaryotes) • • cell walls have peptidoglycan cell walls do not have peptidoglycan Domain Eukarya • four kingdoms of eukaryotes 3 Domains Proteobacteria Domain Archaea Eukaryotes Nanoarchaeota Crenarchaeota Euryarchaeota Korarchaeota Spirochetes Chlamydias Gram-positives Cyanobacteria Gram-positives Epsilon Delta Gamma Beta Alpha Domain Bacteria Domain Eukarya Common ancestor of all living organisms Fig. 24-16, p. 519 Learning Objective 11 • Distinguish among the main groups of archaea based on their ecology • Identify the archaean phyla (Table 24-3) • Describe the main groups of bacteria (Table 24-4) Archaea • Methanogens • • • Extreme halophiles • • produce methane from carbon compounds inhabit anaerobic environments inhabit saturated salt solutions Extreme thermophiles • live at temperatures greater than 100° C Extreme Halophiles Archaeans • • • • Crenarchaeota Euryarchaeota Nanoarchaeota Korarchaeota Archaeans Bacteria • Gram-negative • • • • • Proteobacteria Cyanobacteria Chlamydias Spirochetes Gram-positive bacteria Cyanobacterium Heterocysts 50 µm Fig. 24-18, p. 523 KEY CONCEPTS • • • • Viroids and prions are smaller than viruses A prion consists only of proteins Unlike eukaryotic cells, prokaryotic cells do not have membrane-enclosed organelles such as nuclei and mitochondria Prokaryotes make up two of the three domains: Bacteria and Archaea Learning Objective 12 • What are the ecological roles of prokaryotes, their importance as pathogens, and their commercial importance? Ecological Roles of Prokaryotes • Essential decomposers • recycle nutrients • Some carry out photosynthesis • Many are symbiotic with other organisms Symbiosis • Mutualism • • Commensalism • • • both partners benefit 1 partner benefits other not harmed or helped Parasitism • • parasite benefits host is harmed Bacteria and Disease • Pioneers in microbiology • • • • Anton van Leeuwenhoek Louis Pasteur Robert Koch Koch’s postulates • guidelines to demonstrate specific pathogen causes specific disease symptoms Heliobacter pylori Fig. 24-19a, p. 525 Fig. 24-19b, p. 525 Pathogenic Bacteria • Exotoxins • • strong poisons released by pathogenic bacteria Endotoxins • • poisonous components of cell walls released when bacteria die Pathogenic Bacteria Antibiotic Resistance • Many bacteria have become resistant to antibiotics • R factors • plasmids with genes for antibiotic resistance Commercial Importance • Some bacteria produce antibiotics • Some bacteria used to produce cheese • Lactic acid bacteria used in yogurt, pickles, olives, sauerkraut Lactic Acid Bacteria 5 µm Fig. 24-20, p. 526 KEY CONCEPTS • Great diversity has evolved in the mode of nutrition, the metabolism, and the ecological roles of prokaryotes