Lecture of Cell Signaling-I
advertisement

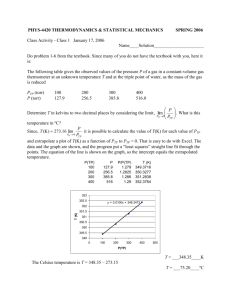
Lecture of Cell Signaling-I Dec. 7, 2004 Contact information: Tzu-Ching Meng Lab 614, IBC, Academia Sinica Tel: 27855696 ext 6140 Email: tcmeng@gate.sinica.edu.tw Phosphorylation is reversible PTPs P P YY Protein P P YY Protein P P PTKs Protein modules in the control of intracellular signaling pathways Docking proteins function as platforms for the recruitment of signaling molecules Models for activation of Signaling proteins A). By membrane translocation B). By conformational change C). By tyrosine phosphorylation Signaling pathways activated by receptor tyrosine kinases Mechanisms for attenuation of receptor tyrosine kinases Classification of human receptor tyrosine kinases (RPTKs) Classification of human cytoplasmic protine tyrosine kinases Activation of receptor tyrosine kinases Juxtamembrane region N-terminal kinase lobe C-terminal tail Substrate precluding loop Substrate accessible loop Activation of c-Src Two modes of intrinsic inhibition by interactions between: (1) SH2 domain and phosphorylated Y527; (2) SH3 domain and Polyproline region. Activation of PKB/Akt PH domain precludes Kinase access by PDK-1 * * * * * In most cases of CML, the leukemic cells share a chromosome abnormality not found in any nonleukemic white blood cells, nor in any other cells of the patient's body. This abnormality is a reciprocal translocation between one chromosome 9 and one chromosome 22. This translocation is designated t(9;22). It results in one chromosome 9 longer than normal and one chromosome 22 shorter than normal. The latter is called the Philadelphia chromosome and designated Ph1. Expression of a fusion PTK p210 Brc-Abl The Protein Tyrosine Phosphatase Superfamily (HCx5R) ‘Classical’ pTyr Specific PTPs (HCSAGxGRxG) Receptor-type PTPs W MAM FN FN SH2 FN FN FN FN FN FN FN FN FN FN FN FN FN VHR-like FN PTEN Cdc25 FN FN FN FN FN FN FN FN FN FN FN FN FN FN FN FN WWW Non-transmembrane PTPs Dual Specificity Phosphatases (HCxxGxxR) C2 FYVE SH2 VHR VH1 PTPH1 MEG1 PTPD1 PTPD2 PEST PTPBAS LyPTP SHP1 MEG2 SHP2 PTP1B TCPTP P E S T PTPb DEP1 SAP1 GLEPP1 CD45 PTPm LAR PTPa PTPg PTPk PTPs PTPe PTPz PTPr PTPd PTPl MKP-1 MKP-2 MKP-3 MKP-4 MKP-5 KAP FYVE- Cdc25A (Cdi1) DSP Cdc25B Cdc25C PTP domain FN SH2 Src Homology domain 2 PEST PEST-like PDZ domain Tonks NK & Neel BG, Curr Opin Cell Biol. 2001, 13(2):182-95 FERM domain Fibronectin III Like repeat Heavily glycosylated Cadherin-like DSP domain Merpin/A5/m MAM domain Immunoglobulin-like W Retinaldehyde Binding protein-like PTEN (MMAC1) Carbonic anhydrase-like FYVE C2 FYVE-domain Lipid binding domain Classification of Protein Tyrosine Phosphatases Non-transmembrane PTPs Receptor-like PTPs Andersen et al., Mol Cell Biol, 21, 7117, 2001 Functional Diversity Through Targeting and Regulatory Domains C-terminal - ER targeting - Proteolytic cleavage Proline rich segment - SH3 binding sites Alternative splicing - Nucleus vs Cytoplasmic SH2 domains - Plasma membrane signaling complexes - Auto-inhibition Cellular retinaldehyde binding protein-like - Golgi targeting - Secretory vesicles - Putative lipid-binding domain FERM domain - Subcellular targeting (e.g. cytoskeletal proteins) PDZ domain(s) - Protein-Protein interactions PEST domain - Protein-Protein Interactions BRO1 domain - Functionally uncharacterised; (Found in a number of signal transduction proteins) - Vesicle associated His-domain - Functionally uncharacterised Sequence comparison of human PTP domains Location of conserved motifs in 3D IVMxT (M6) KCxxYWP (M7) WPDxGxP (M8) TxxD FWxMxW (M5) QTxx QYxF (M10) PxxV HCSAGxGRTG (M9) IAxQGP (M4) NxxKNRY (M1) DYINA (M3) DxxRVxL (M2) http://ptp.cshl.edu Conserved fold of PTP domains N-terminal Central a3-helix Andersen et al Mol. Cell. Biol. 2001 Protein Tyrosine Phosphatase 1B WPD loop PTP Catalytic Mechanism The Protein Tyrosine Phosphatase Superfamily (HCx5R) ‘Classical’ pTyr Specific PTPs (HCSAGxGRxG) Receptor-type PTPs W MAM FN FN SH2 FN FN FN FN FN FN FN FN FN FN FN FN FN VHR-like FN PTEN Cdc25 FN FN FN FN FN FN FN FN FN FN FN FN FN FN FN FN WWW Non-transmembrane PTPs Dual Specificity Phosphatases (HCxxGxxR) C2 FYVE SH2 VHR VH1 PTPH1 MEG1 PTPD1 PTPD2 PEST PTPBAS LyPTP SHP1 MEG2 SHP2 PTP1B TCPTP P E S T PTPb DEP1 SAP1 GLEPP1 CD45 PTPm LAR PTPa PTPg PTPk PTPs PTPe PTPz PTPr PTPd PTPl MKP-1 MKP-2 MKP-3 MKP-4 MKP-5 KAP FYVE- Cdc25A (Cdi1) DSP Cdc25B Cdc25C PTP domain FN SH2 Src Homology domain 2 PEST PEST-like PDZ domain Tonks NK & Neel BG, Curr Opin Cell Biol. 2001, 13(2):182-95 FERM domain Fibronectin III Like repeat Heavily glycosylated Cadherin-like DSP domain Merpin/A5/m MAM domain Immunoglobulin-like W Retinaldehyde Binding protein-like PTEN (MMAC1) Carbonic anhydrase-like FYVE C2 FYVE-domain Lipid binding domain Sequence alignment of amino acid residues at phosphatase motif among human DSPs Amino acid sequence homologies of human DSPs Catalytic mechanism of DSPs Mammalian MAP kinase cascades MAPK and SAPK pathway in mammalian cells T-x-Y at the activation loop Function of MAP Kinase Phosphatases (MKPs) MKP1 MKP2 MKP3 MKP4 MKP5 PAC-1 hVH-5 Pyst2 VHR B23 Localisation Inducible Nuclear Growth factors, Stress Nuclear NGF EGF Cytosolic No Cytosolic Nuclear Cytosolic Cytosolic Nuclear Mitogen No No Stress Mitogen Substrate specificity ERKs=JNKs=p38 ERKs>JNKs>>p38 ERKs>>>JNKs=p38 ERKs>>JNKs=p38 ERK6 ERKs>p38>>JNKs JNKs>p38>>>>ERKs ERK1,ERK2 Mechanism of action of MAP kinase phosphatases (MKPs) Inactivation of MAP kinases (ERK) by threonine or tyrosine dephosphorylation The mammalian MAP kinase phosphatases (MPKs) PTPs and Cancer Refinement of PTP chromosomal positions allows for genetic disease linkage studies 19 PTP chromosomal regions are frequently deleted in human cancers 3 PTP chromosomal regions are frequently duplicated in human cancers PTPs and Cancer PTEN Tumor Suppressor Mutated in various human cancers. Cowden disease DEP1 Tumor suppressor Colon cancer susceptibility locus Scc1 (QTL in mice) PTPk Tumor Suppressor Primary CNS lymphomas SHP2 Noonan Syndrome Developmental disorder affecting 1:2500 newborn Stomach Ulcers Target of Helicobacter pylori Cdc25 Cell Cycle Control Target of Myc and overexpressed in primary breast cancer PRL-3 Metastasis Upregulated in metastases of colon cancer FAP-1 Apoptosis Upregulated in cancers, inhibits CD95-mediated apoptosis PTPs as Drug Targets Immunosupression Diabetes & Obesity Autoimmunity & Allergy PTPs Infectious diseases Cancer Epilepsy Interactions Between PTKs and PTP– (1) PTPs function as NEGATIVE Regulators of Signal Transduction S Autophosphorylation P PTK (Inactive) PTP PTK (Inactive) (Active) S P PTP P P (Active) Interactions Between PTKs and PTPs—(2) PTPs function as POSITIVE Regulators of Signal Transduction PTP P P S S (Inactive) (Active) PTK Important references 1. Hunter, T. (2000) Signaling-2000 and beyond. Cell, 100: 113-127 2. J. Schlessinger (2000) Cell signaling by receptor tyrosine kinases. Cell, 103: 211-225 3. Myers, M. et al. (2001) TYK2 and JAK2 are substrates of protein tyrosine phosphatase 1B. J. Biol. Chem., 276: 47771-47774 4. Andersen, J. N. et al. (2001) Structural and evolutional relationships among protein tyrosine phosphatase domains. Mol. Cell. Biol., 21: 7117-7136 5. Tonks, N. K. (2003) PTP1B: From the sidelines to the front lines. FEBS Letters, 546: 140-148 Additional references 1. Blume-Jensen, P. Hunter, T. (2000) Oncogenic kinase signaling. Cell, 100: 113-127. 2. Palka, H., Park, M. and Tonks, N.K. (2003) Hepatocyte growth factor receptor kinase Met is a substrate of the receptor protein tyrosine phosphatase DEP-1. J. Biol. Chem., 278: 5728-5735. 3. Salmeen, A. et al. (2000) Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B. Mol. Cell, 6: 1404-1412. 4. Meng, T.C. et al (2004) Regulation of insulin signaling through reversible oxidation of the protein-tyrosine phosphatases TC45 and PTP1B. J. Biol. Chem., 279: 37716-37725.