nucleotide_metab_1
advertisement

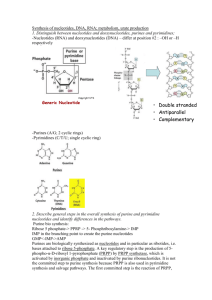
Nucleotides: Synthesis and Degradation Roles of Nucleotides •Precursors to nucleic acids (genetic material and non-protein enzymes). •Currency in energy metabolism (eg. ATP, GTP). •Carriers of activated metabolites for biosynthesis (eg. CDP, UDP). •Structural moieties of coenzymes (eg. NAD, CoA). •Metabolic regulators and signal molecules (eg. cAMP, cGMP, ppGpp). Biosynthetic routes: De novo and salvage pathways De novo pathways Almost all cell types have the ability to synthesize purine and pyrimidine nucleotides from low molecular weight precursors in amounts sufficient for their own needs. The de novo pathways are almost identical in all organisms. Salvage pathways Most organisms have the ability to synthesize nucleotides from nucleosides or bases that become available through the diet or from degredation of nucleic acids. In animals, the extracellular hydrolysis of ingested nucleic acids represents the major route by which bases become available. Reutilization and catabolism of purine and pyrimidine bases blue-catabolism red-salvage pathways endonucleases: pancreatic RNAse pancreatic DNAse phosphodiesterases: usually non-specific PRPP: a central metabolite in de novo and salvage pathways PRPP synthetase Enzyme inhinited by AMP, ADP, and GDP. In E. coli, expression is repressed by PurR repressor bound to either guanine or hypoxanthine. Roles of PRPP: his and trp biosynthesis, nucleobase salvage pathways, de novo synthesis of nucleotides Purine Nucleotide Synthesis O COO OOC 2- O3P O CH2 H O H H H C OH OH OH Aspartate + ATP CH 5 ADP + Pi HC N H SAICAR Synthetase CH2 C COO Ribose Phosphate Pyrophosphokinase AIR Car boxylase AMP ADP + Pi O3P O CH2 H O H H OH OH H O Ribose-5-Phosphate Fumarate P O O O P O C O C H2N CH 5 N C 5-Aminoimidazole Ribotide (AIR) ADP + Pi N10-Formyl- H2C O3P O CH2 H H OH OH HN O C H 2C O C H ADP + Glutamate + Pi FGAM Synthetase NH2 5-Formaminoimidazole-4-carboxamide ribotide (FAICAR) ATP + Glutamine + H2O H 2C IMP Cyclohydrolase O C NH O N10-Formyl-THF THF O HN C O HC C5 NH H H H OH Glycinamide Ribotide (GAR) GAR Transformylase N CH C C N Ribose-5-Phosphate H2O H N CH 5 NH Ribose-5-Phosphate Formylglycinamidine ribotide (FGAM) N C4 NH ADP + Pi OH C H GAR Synthetase AICAR Transformylase THF H2N O -5-Phosphoribosylamine (PRA) Glycine + ATP H CH C NH2 O H O3P O CH2 THF O H N Glutamate + PPi 2- 5-Aminoimidazole-4-carboxamide ribotide (AICAR) ATP Transferase 2- N Ribose-5-Phosphate AIR Synthetase Glutamine + H2O Amidophosphoribosyl CH 5 H2N Ribose-5-Phosphate 5-Phosphoribosyl--pyrophosphate (PRPP) N C4 H2N O Adenylosuccinate Lyase O N HC 4 N 5-Aminoimidazole-4-(N-succinylocarboxamide) ribotide (SAICAR) ATP +HCO3 2- CH 5 H2N Ribose-5-Phosphate Carboxyamidoimidazole Ribotide (CAIR) N C4 N H2N -D-Ribose-5-Phosphate (R5P) ATP C N C4 Ribose-5-Phosphate Formylglycinamide ribotide (FGAR) 4 CH N N 2- O3P O CH2 H H OH O H H OH Inosine Monophosphate (IMP) Example of a salvage pathway: guanine phosphoribosyl transferase In vivo, the reaction is driven to the right by the action of pyrophosphatase Shown: HGPRT, cells also have a APRT. De novo biosynthesis of purines: low molecular weight precursors of the purine ring atoms Synthesis of IMP The base in IMP is called hypoxanthine Note: purine ring built up at nucleotide level. precursors: glutamine (twice) glycine N10-formyl-THF (twice) HCO3 aspartate In vertebrates, 2,3,5 catalyzed by trifunctional enzyme, 6,7 catalyzed by bifunctional enzyme. Pathways from IMP to AMP and GMP G-1: IMP dehydrogenase G-2: XMP aminase A-1: adenylosuccinate synthetase A-2: adenylosuccinate lyase Note: GTP used to make AMP, ATP used to make GMP. Also, feedback inhibition by AMP and GMP. Pathways from AMP and GMP to ATP and GTP Conversion to diphosphate involves specific kinases: GMP + ATP <-------> GDP + ADP Guanylate kinase AMP + ATP <-------> 2 ADP Adenylate kinase Conversion to triphosphate by Nucleoside diphosphate kinase (NDK): GDP + ATP <------> GTP + ADP DG0’= 0 ping pong reaction mechanism with phospho-his intermediate. NDK also works with pyrimidine nucleotides and is driven by mass action. Allosteric regulation of purine de novo synthesis Purine degredation AMP deamination in muscle, hydrolysis in other tissues. Xanthine oxidase:contains FAD, molybdenum, and non-heme iron. In primates, uric acid is the end product, which is excreted. Purine degredation in other animals Clinical disorders of purine metabolism Excessive accumulation of uric acid: Gout The three defects shown each result in elevated de novo purine biosynthesis Common treatment for gout: allopurinol Allopurinol is an analogue of hypoxanthine that strongly inhibits xanthine oxidase. Xanthine and hypoxanthine, which are soluble, are accumulated and excreted. Diseases of purine metabolism (continued) Lesch-Nyhan Syndrome: Severe HGPRT deficiency In addition to symptoms of gout, patients display severe behavioral disorders, learning disorder, aggressiveness and hostility, including selfdirected. Patients must be restrained to prevent self-mutilation. Reason for the behavioral disorder is unknown. X-linked trait (HGPRT gene is on X chromosome). Severe combined immune deficiency (SCID): lack of adenosine deaminase (ADA). Lack of ADA causes accumulation of deoxyadenosine. Immune cells, which have potent salvage pathways, accumulate dATP, which blocks production of other dNTPs by its action on ribonucleotide reductase. Immune cells can’t replicate their DNA, and thus can’t mount an immune response. De novo pyrimidine biosynthesis Pyrimidine ring is assembled as the free base, orotic acid, which is them converted to the nucleotide orotidine monophosphate (OMP). The pathway is unbranched. UTP is a substrate for formation of CTP. Pyrimidine Synthesis O 2 ATP + HCO3- + Glutamine + H2O C 2 ADP + Glutamate + Pi O Carbamoyl Phosphate Synthetase II C C NH2 CH C N H PO3-2 O PRPP C O C C N O HN O CH HN PPi 2- COO O3P O Orotate Phosphoribosyl Transferase CH2 O H H OH OH H H COO Orotidine-5'-monophosphate (OMP) Orotate Carbamoyl Phosphate Aspartate Reduced Quinone Aspartate Transcarbamoylase (ATCase) O O O C C O CH N H CH O 2- CH N H COO COO O3P O CH2 CH N O H2O Dihydroorotase Carbamoyl Aspartate C CH2 HN C C C HN CH2 NH2 CO2 Quinone Pi HO OMP Decarboxylase Dihydroorotate Dehydrogenase O H H OH OH H H Dihydroorotate Uridine Monophosphate (UMP) De novo synthesis of pyrimidines 1: carbamyl phosphate synthase 2: aspartate transcarbamylase 3: dihydroorotase 4: dihydroorotate DH 5: orotate phosphoribosyl tranferase 6: orotidylate decarboxylase 7: UMP kinase 8: NDK 9: CTP synthetase CAD=1,2,3 5 +6=single protein Regulation of pyrimidine de novo synthesis Catabolism of pyrimidines Overview of dNTP biosynthesis One enzyme, ribonucleotide reductase, reduces all four ribonucleotides to their deoxyribo derivitives. A free radical mechanism is involved in the ribonucleotide reductase reaction. There are three classes of ribonucleotide reductase enzymes in nature: Class I: tyrosine radical, uses NDP Class II: adenosylcobalamin. uses NTPs (cyanobacteria, some bacteria, Euglena). Class III: SAM and Fe-S to generate radical, uses NTPs. (anaerobes and fac. anaerobes). Structure of rNDP reductase (E. coli, ClassI) Proposed mechanism for rNDP reductase Proposed reaction mechanism for ribonucleotide reductase Sources of reducing power for rNDP reductase Biological activities of thioredoxin Regulation of activities of mammalian rNDP reductase Salvage and de novo pathways to thymine nucleotides Substrate recvognition by dUTPase Relationship between thymidylate synthase and enzymes of tetrahydrofolate metabolism Catalytic mechanism of thymidylate synthase Regeneration of N5, N10-methylenetetrahydrofolate Biosynthesis of NAD+ and NADP+ Biosynthesis of CoA from pantothenate Proposed reaction mechanism for FGAM synthetase The transformylation reactions are catalyzed by a multiprotein complex components of the complex: GAR transformylase (3) AICAR transformylase (9) serine hydroxymethyl transferase, trifunctional formylmethenylmethylene-THF synthase (activities shown with asterisk) Proposed catalytic mechanism for OMP decarboxylase Reactions catalyzed by eukaryotic dihydroorotate dehydrogenase Nitrogenous Bases Planar, aromatic, and heterocyclic Derived from purine or pyrimidine Numbering of bases is “unprimed” Purines Nitrogenous Bases N1: Aspartate Amine C2, C8: Formate N3, N9: Glutamine C4, C5, N7: Glycine C6: Bicarbonate Ion Pyrimidines Nucleotide Metabolism PURINE RIBONUCLEOTIDES: formed de novo i.e., purines are not initially synthesized as free bases First purine derivative formed is Inosine Mono-phosphate (IMP) The purine base is hypoxanthine AMP and GMP are formed from IMP Purine Nucleotides Get broken down into Uric Acid (a purine) Purine Nucleotide Synthesis ATP is involved in 6 steps and an additional ATP is needed to form the first molecule (R5P) PRPP in the first step of Purine synthesis is also a precursor for Pyrimidine Synthesis, His and Trp synthesis Role of ATP in first step is unique– group transfer rather than coupling In second step, C1 notation changes from to (anomers specifying OH positioning on C1 with respect to C4 group) In step 3, PPi is hydrolyzed to 2Pi (irreversible, “committing” step) Coupling of Reactions Hydrolyzing a phosphate from ATP is relatively easy DG°’= -30.5 kJ/mol If endergonic reaction released energy into cell as heat energy, wouldn’t be useful Must be coupled to an exergonic reaction When ATP is a reactant: or Part of the ATP can be transferred to an acceptor: Pi, PPi, adenyl, adenosinyl group in transferase reaction OR ATP hydrolysis can drive an otherwise unfavorable reaction (synthetase; “energase”) Purine Biosynthetic Pathway Coupling of some reactions on pathway organizes and controls processing of substrates to products in each step Increases overall rate of pathway and protects intermediates from degradation In animals, IMP synthesis pathway is coupled: Reactions 3, 4, 6 Reactions 7, 8 Reactions 10, 11 IMP Conversion to AMP IMP Conversion to GMP Regulatory Control of Purine Nucleotide Biosynthesis GTP is involved in AMP synthesis and ATP is involved in GMP synthesis (reciprocal control of production) PRPP is a biosynthetically “central” molecule (why?) ADP/GDP levels – negative feedback on Ribose Phosphate Pyrophosphokinase Amidophosphoribosyl transferase is activated by PRPP levels APRT activity has negative feedback at two sites ATP, ADP, AMP bound at one site GTP,GDP AND GMP bound at the other site Rate of AMP production increases with increasing concentrations of GTP; rate of GMP production increases with increasing concentrations of ATP Regulatory Control of Purine Biosynthesis At level of IMP production: Independent control Synergistic control Feedforward activation by PRPP Below level of IMP production Reciprocal control Total amounts of purine nucleotides controlled Relative amounts of ATP, GTP controlled Purine Catabolism and Salvage All purine degradation in animals leads to uric acid Ingested nucleic acids are degraded by pancreatic nucleases, and intestinal phosphodiesterases in the intestine Group-specific nucleotidases and non-specific phosphatases degrade nucleotides into nucleosides Direct absorption of nucleosides Further degradation Nucleoside + H2O base + ribose (nucleosidase) Nucleoside + Pi base + r-1-phosphate (n. phosphorylase) NOTE: MOST INGESTED NUCLEIC ACIDS ARE DEGRADED AND EXCRETED. Intracellular Purine Catabolism Nucleotides broken into nucleosides by action of 5’nucleotidase (hydrolysis reactions) Purine nucleoside phosphorylase (PNP) Inosine Hypoxanthine Xanthosine Xanthine Guanosine Guanine Ribose-1-phosphate splits off Can be isomerized to ribose-5-phosphate Adenosine is deaminated to Inosine (ADA) Intracellular Purine Catabolism Xanthine is the point of convergence for the metabolism of the purine bases Xanthine Uric acid Xanthine oxidase catalyzes two reactions Purine ribonucleotide degradation pathway is same for purine deoxyribonucleotides Adenosine Degradation Xanthosine Degradation • Ribose sugar gets recycled (Ribose-1-Phosphate R-5-P ) – can be incorporated into PRPP (efficiency) • Hypoxanthine is converted to Xanthine by Xanthine Oxidase • Guanine is converted to Xanthine by Guanine Deaminase • Xanthine gets converted to Uric Acid by Xanthine Oxidase Xanthine Oxidase A homodimeric protein Contains electron transfer proteins FAD Mo-pterin complex in +4 or +6 state Two 2Fe-2S clusters Transfers electrons to O2 H2O2 H2O2 is toxic Disproportionated to H2O and O2 by catalase THE PURINE NUCLEOTIDE CYCLE AMP + H2O IMP + NH4+ (AMP Deaminase) IMP + Aspartate + GTP AMP + Fumarate + GDP + Pi (Adenylosuccinate Synthetase) COMBINE THE TWO REACTIONS: Aspartate + H2O + GTP Fumarate + GDP + Pi + NH4+ The overall result of combining reactions is deamination of Aspartate to Fumarate at the expense of a GTP Purine Nucleotide Cycle In-Class Question: Why is the purine nucleotide cycle important in muscle metabolism during a burst of activity? Adenosine Deaminase CHIME Exercise: 2ADA Enzyme catalyzing deamination of Adenosine to Inosine / barrel domain structure “TIM Barrel” – central barrel structure with 8 twisted parallel -strands connected by 8 -helical loops Active site is at bottom of funnel-shaped pocket formed by loops Found in all glycolytic enzymes Found in proteins that bind and transport metabolites Uric Acid Excretion Humans – excreted into urine as insoluble crystals Birds, terrestrial reptiles, some insects – excrete isoluble crystals in paste form (conserve water) Others – further modification : Uric Acid Allantoin Allantoic Acid Urea Ammonia Purine Salvage Adenine phosphoribosyl transferase (APRT) Adenine + PRPP AMP + PPi Hypoxanthine-Guanine phosphoribosyl transferase (HGPRT) Hypoxanthine + PRPP IMP + PPi Guanine + PRPP GMP + PPi (NOTE: THESE ARE ALL REVERSIBLE REACTIONS) AMP,IMP,GMP do not need to be resynthesized de novo ! A CASE STUDY : GOUT A 45 YEAR OLD MAN AWOKE FROM SLEEP WITH A PAINFUL AND SWOLLEN RIGHT GREAT TOE. ON THE PREVIOUS NIGHT HE HAD EATEN A MEAL OF FRIED LIVER AND ONIONS, AFTER WHICH HE MET WITH HIS POKER GROUP AND DRANK A NUMBER OF BEERS. HE SAW HIS DOCTOR THAT MORNING, “GOUTY ARTHRITIS” WAS DIAGNOSED, AND SOME TESTS WERE ORDERED. HIS SERUM URIC ACID LEVEL WAS ELEVATED AT 8.0 mg/dL (NL < 7.0 mg/dL). THE MAN RECALLED THAT HIS FATHER AND HIS GRANDFATHER, BOTH OF WHOM WERE ALCOHOLICS, OFTEN COMPLAINED OF JOINT PAIN AND SWELLING IN THEIR FEET. A CASE STUDY : GOUT THE DOCTOR RECOMMENDED THAT THE MAN USE NSAIDS FOR PAIN AND SWELLING, INCREASE HIS FLUID INTAKE (BUT NOT WITH ALCOHOL) AND REST AND ELEVATE HIS FOOT. HE ALSO PRESCRIBED ALLOPURINOL. A FEW DAYS LATER THE CONDITION HAD RESOLVED AND ALLOPURINOL HAD BEEN STOPPED. A REPEAT URIC ACID LEVEL WAS OBTAINED (7.1 mg/dL). THE DOCTOR GAVE THE MAN SOME ADVICE REGARDING LIFE STYLE CHANGES. Gout Impaired excretion or overproduction of uric acid Uric acid crystals precipitate into joints (Gouty Arthritis), kidneys, ureters (stones) Lead impairs uric acid excretion – lead poisoning from pewter drinking goblets Fall of Roman Empire? Xanthine oxidase inhibitors inhibit production of uric acid, and treat gout Allopurinol treatment – hypoxanthine analog that binds to Xanthine Oxidase to decrease uric acid production ALLOPURINOL IS A XANTHINE OXIDASE INHIBITOR A SUBSTRATE ANALOG IS CONVERTED TO AN INHIBITOR, IN THIS CASE A “SUICIDE-INHIBITOR” Lesch-Nyhan Syndrome A defect in production or activity of HGPRT Causes increased level of Hypoxanthine and Guanine ( in degradation to uric acid) Also,PRPP accumulates stimulates production of purine nucleotides (and thereby increases their degradation) Causes gout-like symptoms, but also neurological symptoms spasticity, aggressiveness, self-mutilation First neuropsychiatric abnormality that was attributed to a single enzyme Purine Autism 25% of autistic patients may overproduce purines To diagnose, must test urine over 24 hours Biochemical findings from this test disappear in adolescence Must obtain urine specimen in infancy, but it’s difficult to do! Pink urine due to uric acid crystals may be seen in diapers Pyrimidine Ribonucleotide Synthesis Uridine Monophosphate (UMP) is synthesized first CTP is synthesized from UMP Pyrimidine ring synthesis completed first; then attached to ribose-5-phosphate N1, C4, C5, C6 : Aspartate C2 : HCO3N3 : Glutamine amide Nitrogen UMP Synthesis Overview 2 ATPs needed: both used in first step One transfers phosphate, the other is hydrolyzed to ADP and Pi 2 condensation rxns: form carbamoyl aspartate and dihydroorotate (intramolecular) Dihydroorotate dehydrogenase is an intra-mitochondrial enzyme; oxidizing power comes from quinone reduction Attachment of base to ribose ring is catalyzed by OPRT; PRPP provides ribose-5-P PPi splits off PRPP – irreversible Channeling: enzymes 1, 2, and 3 on same chain; 5 and 6 on same chain Pyrimidine Synthesis O 2 ATP + HCO3- + Glutamine + H2O C 2 ADP + Glutamate + Pi O Carbamoyl Phosphate Synthetase II C C NH2 CH C N H PO3-2 O PRPP C O C C N O HN O CH HN PPi 2- COO O3P O Orotate Phosphoribosyl Transferase CH2 O H H OH OH H H COO Orotidine-5'-monophosphate (OMP) Orotate Carbamoyl Phosphate Aspartate Reduced Quinone Aspartate Transcarbamoylase (ATCase) O O O C C O CH N H CH O 2- CH N H COO COO O3P O CH2 CH N O H2O Dihydroorotase Carbamoyl Aspartate C CH2 HN C C C HN CH2 NH2 CO2 Quinone Pi HO OMP Decarboxylase Dihydroorotate Dehydrogenase O H H OH OH H H Dihydroorotate Uridine Monophosphate (UMP) UMP UTP and CTP Nucleoside monophosphate kinase catalyzes transfer of Pi to UMP to form UDP; nucleoside diphosphate kinase catalyzes transfer of Pi from ATP to UDP to form UTP CTP formed from UTP via CTP Synthetase driven by ATP hydrolysis Glutamine provides amide nitrogen for C4 OMP DECARBOXYLASE : THE MOST CATALYTICALLY PROFICIENT ENZYME FINAL REACTION OF PYRIMIDINE PATHWAY ANOTHER MECHANISM FOR DECARBOXYLATION A CARBANION INTERMEDIATE (UNSTABLE) MUST BE STABILIZED BUT NO COFACTORS ARE NEEDED! SOME OF THE BINDING ENERGY BETWEEN OMP AND THE ACTIVE SITE IS USED TO STABILIZE THE TRANSITION STATE “PREFERENTIAL TRANSITION STATE BINDING” Regulatory Control of Pyrimidine Synthesis Differs between bacteria and animals Bacteria – regulation at ATCase rxn Animals – regulation at carbamoyl phosphate synthetase II UDP and UTP inhibit enzyme; ATP and PRPP activate it UMP and CMP competitively inhibit OMP Decarboxylase *Purine synthesis inhibited by ADP and GDP at ribose phosphate pyrophosphokinase step, controlling level of PRPP also regulates pyrimidines Orotic Aciduria Caused by defect in protein chain with enzyme activities of last two steps of pyrimidine synthesis Increased excretion of orotic acid in urine Symptoms: retarded growth; severe anemia Only known inherited defect in this pathway (all others would be lethal to fetus) Treat with uridine/cytidine IN-CLASS QUESTION: HOW DOES URIDINE AND CYTIDINE ADMINISTRATION WORK TO TREAT OROTICACIDURIA? Degradation of Pyrimidines CMP and UMP degraded to bases similarly to purines Dephosphorylation Deamination Glycosidic bond cleavage Uracil reduced in liver, forming -alanine Converted to malonyl-CoA fatty acid synthesis for energy metabolism Deoxyribonucleotide Formation Purine/Pyrimidine degradation are the same for ribonucleotides and deoxyribonucleotides Biosynthetic pathways are only for ribonucleotides Deoxyribonucleotides are synthesized from corresponding ribonucleotides DNA vs. RNA: REVIEW DNA composed of deoxyribonucleotides Ribose sugar in DNA lacks hydroxyl group at 2’ Carbon Uracil doesn’t (normally) appear in DNA Thymine (5-methyluracil) appears instead Formation of Deoxyribonucleotides Reduction of 2’ carbon done via a free radical mechanism catalyzed by “Ribonucleotide Reductases” E. coli RNR reduces ribonucleoside diphosphates (NDPs) to deoxyribonucleoside diphosphates (dNDPs) Two subunits: R1 and R2 A Heterotetramer: (R1)2 and (R2)2 in vitro RIBONUCLEOTIDE REDUCTASE R1 SUBUNIT Specificity Site Hexamerization site Activity Site Five redox-active –SH groups from cysteines R2 SUBUNIT Tyr 122 radical Binuclear Fe(III) complex Chime Exercise E. coli Ribonucleotide Reductase: 3R1R and 4R1R: R1 subunit 1RIB and 1AV8: R2 subunit Ribonucleotide Reductase R2 Subunit Fe prosthetic group– binuclear, with each Fe octahedrally coordinated Fe’s are bridged by O-2 and carboxyl gp of Glu 115 Tyr 122 is close to the Fe(III) complex stabilization of a tyrosyl free-radical During the overall process, a pair of –SH groups provide the reducing equivalents A protein disulfide group is formed Gets reduced by two other sulfhydryl gps of Cys residues in R1 Mechanism of Ribonucleotide Reductase Reaction Free Radical Involvement of multiple –SH groups RR is left with a disulfide group that must be reduced to return to the original enzyme RIBONUCLEOTIDE REDUCTASE ACTIVITY IS RESPONSIVE TO LEVEL OF CELLULAR NUCLEOTIDES: ATP ACTIVATES REDUCTION OF CDP UDP dTTP INDUCES GDP REDUCTION INHIBITS REDUCTION OF CDP. UDP dATP INHIBITS REDUCTION OF ALL NUCLEOTIDES dGTP STIMULATES ADP REDUCTION INHIBITS CDP,UDP,GDP REDUCTION RIBONUCLEOTIDE REDUCTASE CATALYTIC ACTIVITY VARIES WITH STATE OF OLIGOMERIZATION: WHEN ATP, dATP, dGTP, dTTP BIND TO SPECIFICITY SITE OF R1 (CATALYTICALLY INACTIVE MONOMER) CATALYTICALLY ACTIVE (R1)2 WHEN dATP OR ATP BIND TO ACTIVITY SITE OF DIMERS (R1)4a TETRAMER FORMATION (ACTIVE STATE) == (R1)4b (INACTIVE) WHEN ATP BINDS TO HEXAMERIZATION SITE CATALYTICALLY ACTIVE HEXAMERS (R1)6 Thioredoxin Physiologic reducing agent of RNR Cys pair can swap H atoms with disulfide formed regenerate original enzyme Thioredoxin gets oxidized to disulfide Oxidized Thioredoxin gets reduced by thioredoxin reductase mediated by NADPH (final electron acceptor) Thymine Formation Formed by methylating deoxyuridine monophosphate (dUMP) UTP needed for RNA production, but dUTP not needed for DNA If dUTP produced excessively, would cause substitution errors (dUTP for dTTP) dUTP hydrolyzed by dUTP diphosphohydrolase to dUMP methylated at C5 to form dTMP rephosphorylate to form dTTP Chime Exercise 1DUD: dUTPase Tetrahydrofolate (THF) Methylation of dUMP catalyzed by thymidylate synthase Cofactor: N5,N10-methylene THF Oxidized to dihydrofolate Only known rxn where net oxidation state of THF changes THF Regeneration: DHF + NADPH + H+ THF + NADP+ (enzyme: dihydrofolate reductase) THF + Serine N5,N10-methylene-THF + Glycine (enzyme: serine hydroxymethyl transferase) Anti-Folate Drugs Cancer cells consume dTMP quickly for DNA replication Interfere with thymidylate synthase rxn to decrease dTMP production (fluorodeoxyuridylate – irreversible inhibitor) – also affects rapidly growing normal cells (hair follicles, bone marrow, immune system, intestinal mucosa) Dihydrofolate reductase step can be stopped competitively (DHF analogs) Anti-Folates: Aminopterin, methotrexate, trimethoprim IN-CLASS QUESTION IN von GIERKE’S DISEASE, OVERPRODUCTION OF URIC ACID OCCURS. THIS DISEASE IS CAUSED BY A DEFICIENCY OF GLUCOSE-6-PHOSPHATASE. EXPLAIN THE BIOCHEMICAL EVENTS THAT LEAD TO INCREASED URIC ACID PRODUCTION? WHY DOES HYPOGLYCEMIA OCCUR IN THIS DISEASE? WHY IS THE LIVER ENLARGED? ADENOSINE DEAMINASE DEFICIENCY IN PURINE DEGRADATION, ADENOSINE INOSINE ENZYME IS ADA ADA DEFICIENCY RESULTS IN SCID “SEVERE COMBINED IMMUNODEFICIENCY” SELECTIVELY KILLS LYMPHOCYTES BOTH B- AND T-CELLS MEDIATE MUCH OF IMMUNE RESPONSE ALL KNOWN ADA MUTANTS STRUCTURALLY PERTURB ACTIVE SITE ADA DEFICIENCY IN-CLASS QUESTION: EXPLAIN THE BIOCHEMISTRY THAT RESULTS WHEN A PERSON HAS ADA DEFICIENCY (HINT: LYMPHOID TISSUE IS VERY ACTIVE IN DEOXYADENOSINE PHOSPHORYLATION) ADA DEFICIENCY ONE OF FIRST DISEASES TO BE TREATED WITH GENE THERAPY ADA GENE INSERTED INTO LYMPHOCYTES; THEN LYMPHOCYTES RETURNED TO PATIENT PEG-ADA TREATMENTS ACTIVITY LASTS 1-2 WEEKS Sugars Pentoses (5-C sugars) Numbering of sugars is “primed” Sugars D-Ribose and 2’-Deoxyribose *Lacks a 2’-OH group Nucleosides Result from linking one of the sugars with a purine or pyrimidine base through an Nglycosidic linkage Purines bond to the C1’ carbon of the sugar at their N9 atoms Pyrimidines bond to the C1’ carbon of the sugar at their N1 atoms Nucleosides Phosphate Groups Mono-, di- or triphosphates Phosphates can be bonded to either C3 or C5 atoms of the sugar Nucleotides Result from linking one or more phosphates with a nucleoside onto the 5’ end of the molecule through esterification Nucleotides RNA (ribonucleic acid) is a polymer of ribonucleotides DNA (deoxyribonucleic acid) is a polymer of deoxyribonucleotides Both deoxy- and ribonucleotides contain Adenine, Guanine and Cytosine Ribonucleotides contain Uracil Deoxyribonucleotides contain Thymine Nucleotides Monomers for nucleic acid polymers Nucleoside Triphosphates are important energy carriers (ATP, GTP) Important components of coenzymes FAD, NAD+ and Coenzyme A Naming Conventions Nucleosides: Purine nucleosides end in “-sine” Adenosine, Cytosine Pyrimidine nucleosides end in “-dine” Thymidine, Guanidine Nucleotides: Start with the nucleoside name from above and add “mono-”, “di-”, or “triphosphate” Adenosine Monophosphate, Guanidine Triphosphate, Deoxythymidine Diphosphate In-Class Activities Look at the Nucleotide Structures Take the Nucleotide Identification Quiz Be prepared to identify some of these structures on an exam. Learn some “tricks” that help you to distinguish among the different structures