Homework 10 ans key
advertisement

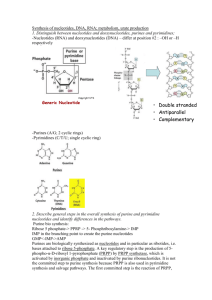
FCH 532 Study Guide This assignment will not be collected 1. Starting out from ribose-5-phosphate, write out the metabolic pathway for the production of inosine monophosphate (IMP). Show all chemical structures and names of substrates. Also show co-factors and enzymes. a. Continue from IMP to produce AMP and GMP. How is purine synthesis regulated? 2. Show the pathways for the de novo synthesis of pyrimidines. In what ways does this differ from purine synthesis. 3 possible pyrimidines U, C, and T. Pyrimidines are simpler to synthesize than purines. Fewer steps. N1, C4, C5, and C6 come from Asp C2 from bicarbonate N3 from Gln Start by synthesizing uracil monophosphate (UMP) After UMP is formed , converted to UTp by nucleoside monophosphate kinase and nucleoside diphosphate kinase respectively. UMP + ATP UDP + ADP UDP + ATP UTP + ADP UTP is converted to CTP by CTP synthetase. CTP is formed by amination of UTP by CTP synthetase In animals, amino group from Gln T is made from dUMP and is more complicated. •2 main enzymes: dUTP diphosphohydrolase (dUTPase) and thymidylate synthase Reaction 1 •dTMP is made by methylation of dUMP. •dUMP is made by hydrolysis of dUTP via dUTP diphosphohydrolase (dUTPase) dUTP + H2O dUMP+ PPi •Done to minimize the concentration of dUTP-prevents incorporation of uracil into DNA. Reaction 2 •dTMP is made from dUMP by thymidylate synthase (TS). Uses N5, N10-methylene-THF as methyl donor In bacteria, amino group from ammonia 3. How is pyrimidine biosynthesis regulated in E. coli as compared to in animals? 4. What is the function of ribonucleotide reductase? Write out a mechanism for this enzyme’s reaction. RNR is necessary to produce deoxyribose derivatives by reducing ribonucleotides at the C2’ position. 5. Write out a plausible mechanism for the inhibition of thymidylate synthetase by FdUMP. H2N N H N O N+ CH2 R CH2 N HN O- O F HN O :B H -S E dRib-P N CH2 H F HN H S N O :B E dRib-P H2N N H N CH2 H+ H N CH2 O R O CH2 N F HN H :B N O S E dRib-P HN 6. Write out the major catabolic pathways for purines and pyrimidines. Show all chemical structures. See above and notes. 7. Write the pathway for the biosynthesis of uridine triphosphate UTP) naming all enzymes, coenzymes, and substrates. You should show the structure of all substrates but need not show the structures of the enzyme intermediates See above. 8. Briefly discuss the biochemical mechanism by which a cell regulates purine biosynthesis so it obtains balanced amounts of both purine nucleotides. In order to achieve balanced amounts of purine nucleotides, the reactions catalyze the first steps that convert IMP to either AMP or GMP are regulated by feedback inhibition. Adenylosuccinate synthase which converts IMP to adenylosuccinate is inhibited by AMP and ADP. IMP dehydrogenase that converts IMP to xanthosine monophosphate (XMP) is inhibited by GMP and GDP. ATP activates the GMP synthetase which converts XMP to GMP and GTP activates adenylosuccinate synthase. 9. Most nucleotide bases are reused rather than synthesized from scratch. Briefly discuss the reutilization pathways for purines and contrast the difference in ATP requirements to reutilize adenine for the synthesis of ATP versus forming a new ATP by de novo synthesis. Free purines such as adenine, guanine, and hypoxanthine can be reconverted to their corresponding nucleotides through salvage pathways. These are catalyzed by two enzymes: Adeninephosphoribosyltransferase (APRT) Adenine + PRPP AMP + PPi and Hypoxanthine-guanine phosphoribosyltransferase (HGPRT) Hypoxanthine + PRPP IMP + PPi Guanine + PRPP GMP + PPi These require far less energy than synthesizing these molecules from purine biosynthesis which uses additional 5 ATP molecules vs. the 1 necessary for regenerating PRPP (purine biosynthesis also uses this ATP to generate PRPP, thus consuming 6 ATP molecules per nucleotide formed). 10. Inhibitors of one-carbon metabolism (THF) block the synthesis of DNA but not RNA, why? N5, N10-Methylenetetrahydrofolate (THF) is an essential cofactor for thymidylate synthase, the enzyme responsible for the production of dTMP. If THF metabolism is inhibited, dTMP production is inhibited. Since thymine is essential for DNA synthesis but not RNA synthesis, only DNA synthesis will be inhibited. 11. Name the different nucleotides, their nucleosides and their component bases. 12. Define the following terms: Z-scheme, photorespiration, photosystem 2, light harvesting complex, chlorophyll, Calvin cycle, lumen, photoautotroph, Rubisco, etc. Z-scheme: describes two-center electron transport. This system uses reding power derived from light energy to drive the oxidation of water to produce NADPH. It requires two processes: The first catalyzed by Photosystem II (PSII) that generates a strong oxidant to oxidize H2O and a weak reductant. The second catalyzed by Photosystem I (PSI) that generates a strong reductant capable of reducing NADP+. photorespiration: Illuminated plants can consume O2 and evolve CO2 distinct from oxidative phosphorylation. O2 competes with CO2 as substrate for RUBP carboxylase. O2 reacts with RuBP to form 3PG and 2-phosphoglycolate. 2-phosphoglycolate is hydrolyzed to glycolate by glycolate phosphatase. Partially oxidized to yield CO2 in peroxisome(aka glyoxisome) and mitochondria. Dissipates some ATP and NADPH generated by light reactions. photosystem 2: AKA photosystem II (PSII). An oxygen evolving complex responsible for the oxidation of of water. Two molecules of ChlA make up P680 is the photon absorbing center. Absorbance of a photon converts P680 to P680* which then donates electron to Pheo a which then passes it’s electron to quinones QA and QB and to the quinone pool. producing P680+ which can return to the original state by removing electron from TyrZ. TyrZ derives its electrons from the oxygen evolving complex (OEX) which contains 4 Mn++ ions. It takes 4 photons of light to charge P680+ 4 times to take 2H2O to O2 and 4H+. light harvesting complex (LHC): Divided into two types in purple photosynthetic bacteria (LH1 and LH2) are transmembrane proteins that have different spectral and biochemical properties. LH2 absorbes light at shorter wavelengths than LH1. LH2 passes energy from photons it absorbs to LH1 which passes them to the reaction center. LH2 binds 24 bacteriochlorphyll a and 8 lycopene molecules. Many are also associated with accessory pigments that fill the absorption spectra where chlophylls do not absorb strongly. antenna chlorophyll: Because reaction centers (RCs) can only intercept ~1 photon per second, antenna chlorophyll perform an essential role in increasing the efficiency of photosynthesis. These antenna can pass the energy of an aborbed photon by exciton transfer from molecule to molecule until they reach a reaction center. These LHCs can transfer the energy to an RC in <10-10 s with an efficiencty of >90%. Chlorophyll: Primary photoreceptor for photosynthesis is a cyclic tetrapyrrole that differs from heme in four major ways. 1.Central metal ion is Mg2+ not Fe(II) or Fe(III). 2. Has cyclopentenone ring, (Ring V), fused to pyrrole Ring III 3. Pyrolle Ring IV is partially reduced in chlorophyll a (Chl a) and chlorophyll b (Chl b). In bacteriochlorophyll Rings II and IV are partially reduced. 4. Propionyl side chain of Ring IV is esterified to tetraisoprenoid alcohol. In Chl a and b and Bchlb it is phytol. Calvin cycle: aka reductive pentose phosphate cycle generates GAP from CO2 Lumen: space inside the thylakoid compartment. Photoautotroph: An organism, that can use energy from sunlight to convert CO2 into carbohydrate for use in cellular functions. Rubisco: ribulose-1,5-bisphosphate carboxylase oxygenase enzyme that catalyzes the first carbon fixation step in the Calvin cycle. 3. One of the classic experiments conducted by Benson, Bassham and Calvin as part of their elucidation of the pathway of dark carbon fixation was to feed the algae 14 -C labeled carbon dioxide and then degrade the resulting sugars to determine which carbons are radio-labeled. Indicate which carbons will be radiolabelled in the following sugars after feeding radioactive carbon dioxide. See above (red carbons in Figure 24-31) 4. What are the 4 possible routes of dissipation of excitation energy during phototransfer? Internal conversion-electronic energy is converted to heat (molecular motion). Occurs very rapidly (<10-11s) and molecules returned to ground state. Fluorescence-electronic energy is reduced to ground state by emitting a photon Occurs slower than internal conversion (~10-8s). Emitted photon has a longer wavelength (lower energy) than the initially absorbed photon. Accounts for 3-6% of light energy absorbed-usually causes red fluorescence. Exciton transfer (resonance energy transfer)-electronic energy is directly transferred to nearby unexcited molecules with similar electronic properties Funnels the light to photosynthetic reaction centers Photooxidation-light-excited donor molecule is oxidized by transferring an electron to an acceptor molecule.