Modification of Amino Acids
advertisement

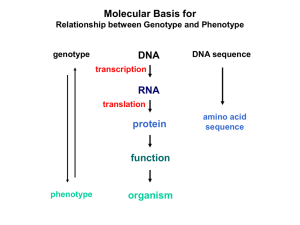
Colinearity of Gene and Protein genotype DNA DNA sequence transcription RNA translation protein function phenotype organism amino acid sequence Colinearity of Gene and Protein “The linear sequence of nucleotides in a gene determines the linear sequence of amino acids in a protein.” Mutant alleles of trpA gene differed in the position of the mutation at the DNA level, which corresponded to position of amino acid substitution in the gene product. Colinearity of mutations and altered amino acids in a subunit of tryptophan synthetase from E. coli C. Yanofsky, 1967. Scientific American Molecular Basis for Relationship between Genotype and Phenotype genotype DNA DNA sequence transcription RNA translation protein function phenotype organism amino acid sequence tRNA Anticodon of a tRNA molecule recognizes and pairs with an mRNA codon. tRNA contains modified bases: pseudouridine, methylguanosine, dimethylguanosine, methylinosine, dihydrouridine. Genetic Code Aminoacyl-tRNA Synthetase Attaches Amino Acid to tRNA Aminoacyl-tRNA synthetase catalyzes the formation of “charged” tRNA. There is an aminoacyltRNA synthetase for each amino acid. The carboxyl end of an amino acid is attached to the 3’ end of the tRNA. Wobble Position Some tRNA molecules can recognize and pair with more than one specific codon. Base-pairing between the 3’ base of a codon and 5’ base of an anticodon is not always exact. Molecular Basis for Relationship between Genotype and Phenotype genotype DNA DNA sequence transcription RNA translation protein function phenotype organism amino acid sequence Protein Synthesis: Brief Summary 3 Stages • Initiation • Elongation • Termination Catalytic Proteins • Initiation Factors • Elongation Factors • Termination Factors Hydrolysis of GTP provides energy to drive some reactions. mRNA, rRNA, and tRNA are involved. Protein Synthesis: Initiation in Prokaryotes Shine-Dalgarno sequence pairs with 16S rRNA of 30S subunit. IF3 keeps 30S subunit dissociated from 50S subunit. Formyl group is added to methionine when associated with the initiator tRNA. IF1 and IF2 allows only initiator tRNA to enter P site. Initiation factors are released when two ribosomal subunits associate. Protein Synthesis: Initiation in Eukaryotes eIF4A, eIF4B, and eIF4G associates with 5’ end, then with 40S subunit and initiator tRNA. mRNA is unwound by movement of this complex in 5’ -> 3’ direction. 60S subunit associates with initiation complex when start codon is recognized. Initiation factors are released when the two ribosomal subunits associate. Important Features of Ribosome A - aminoacyl site P - peptidyl site E - exit site Protein Synthesis: Elongation EF-Tu associates with aminoacyl-tRNA to form a ternary complex. Correct match of ternary complex with codon in A site (decoding center) changes conformation of ribosome. EF-Tu leaves ternary complex, and peptide bond is formed between amino acids as amino acids are positioned together in peptidyltransferase center. Amino acid in P site is transferred to amino acid in A site. Translocation requires GTP and EF-G. EF-G enters A site, shifting tRNAs. When EF-G leaves, A site is open for a new ternary complex. A new ternary complex associates with A site, and deacylated tRNA leaves from E site. Protein Synthesis: Termination tRNA molecules do not recognize stop codons. Termination codons are recognized by release factors. (RF1, RF2, RF3 in bacteria) UAA and UAG are recognized by RF1. UAA and UGA are recognized by RF2. RF3 assists in release activity. Release factors bind to a stop codon in the A site by association between codon and tripeptide of RF. Polypeptide is released from P site when RF fits into A site. Release of polypeptide is followed by dissociation of ribosomal subunits. Molecular Basis for Relationship between Genotype and Phenotype genotype DNA DNA sequence transcription RNA translation protein function phenotype organism amino acid sequence Molecular Basis for Relationship between Genotype and Phenotype genotype DNA DNA sequence transcription RNA translation protein function phenotype organism amino acid sequence All Protein Interactions in an Organism Compose the Interactome Proteome: Complete set of proteins produced by genetic material of an organism. Interactome: Complete set of protein interactions in an organism. Alternative Splicing Produces Related but Distinct Protein Isoforms Posttranslational Events Protein Folding: Translational product (polypeptide) achieves appropriate folding by aid of chaperone proteins. Modification of Amino Acids: * Phosphorylation/dephosphorylation * Ubiquitination Protein Targeting: Directing proteins to specific locations (for example, nucleus, mitochondria, or cell membrane) is accomplished by tagging of proteins (signal sequence for secreted proteins, nuclear localization sequences for nuclear proteins). Posttranslational Events Protein Folding: Translational product (polypeptide) achieves appropriate folding by aid of chaperone proteins. Modification of Amino Acids: * Phosphorylation/dephosphorylation * Ubiquitination Protein Targeting: Directing proteins to specific locations (for example, nucleus, mitochondria, or cell membrane) is accomplished by tagging of proteins (signal sequence for secreted proteins, nuclear localization sequences for nuclear proteins). Phosphorylation and Dephosphorylation of Proteins Kinases add phosphate groups to hydroxyl groups of amino acids such as serine and threonine. Phosphatases remove phosphate groups. Ubiquitinization Targets a Protein for Degradation Short-lived proteins are ubiquitinated: • cell-cycle regulators • damaged proteins Posttranslational Events Protein Folding: Translational product (polypeptide) achieves appropriate folding by aid of chaperone proteins. Modification of Amino Acids: * Phosphorylation/dephosphorylation * Ubiquitination Protein Targeting: Directing proteins to specific locations (for example, nucleus, mitochondria, or cell membrane) is accomplished by tagging of proteins (signal sequence for secreted proteins, nuclear localization sequences for nuclear proteins). Signal Sequences Target Proteins for Secretion Signal sequence at the amino-terminal end of membrane proteins or secretory proteins are recognized by factors and receptors that mediate transmembrane transport. Signal sequence is cleaved by signal peptidase. Nuclear localization sequences (NLSs) are located in interior of proteins such as DNA and RNA polymerases. They are recognized by nuclear pore proteins for transport into nucleus. Universality of Genetic Information Transfer Genetic code is essentially identical for all organisms. There are exceptions. System “universal” mammalian mitochondria yeast mitochondria AUA isoleucine methionine isoleucine UGA termination tryptophan tryptophan Comparison of Gene Expression Prokaryotes Eukaryotes One type of RNA polymerase synthesizes all RNA molecules. Three different types of RNA polymerases synthesize different classes of RNA. mRNA is translated during transcription. mRNA is processed before translation. Genes are not split. They are continguous segments of DNA. Genes are often split. They are not continguous segments of coding sequences. mRNAs are often polycistronic. mRNAs are mostly monocistronic.