1 - CSUMB iLearn
advertisement
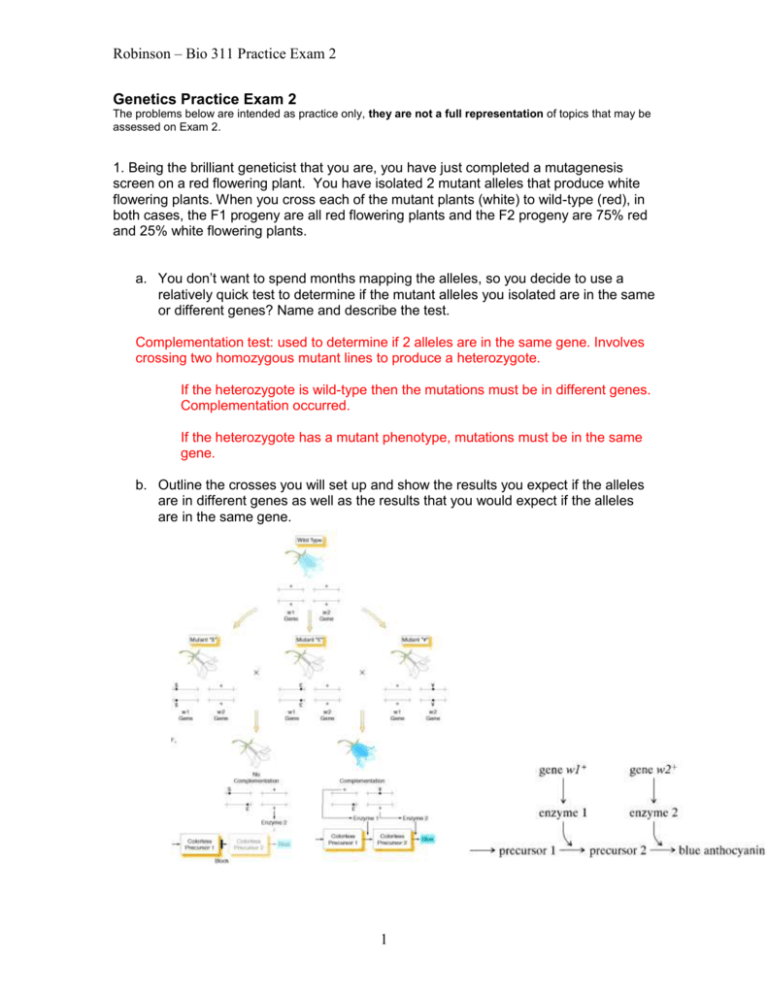
Robinson – Bio 311 Practice Exam 2 Genetics Practice Exam 2 The problems below are intended as practice only, they are not a full representation of topics that may be assessed on Exam 2. 1. Being the brilliant geneticist that you are, you have just completed a mutagenesis screen on a red flowering plant. You have isolated 2 mutant alleles that produce white flowering plants. When you cross each of the mutant plants (white) to wild-type (red), in both cases, the F1 progeny are all red flowering plants and the F2 progeny are 75% red and 25% white flowering plants. a. You don’t want to spend months mapping the alleles, so you decide to use a relatively quick test to determine if the mutant alleles you isolated are in the same or different genes? Name and describe the test. Complementation test: used to determine if 2 alleles are in the same gene. Involves crossing two homozygous mutant lines to produce a heterozygote. If the heterozygote is wild-type then the mutations must be in different genes. Complementation occurred. If the heterozygote has a mutant phenotype, mutations must be in the same gene. b. Outline the crosses you will set up and show the results you expect if the alleles are in different genes as well as the results that you would expect if the alleles are in the same gene. 1 Robinson – Bio 311 Practice Exam 2 2. In corn, the alleles c+ and c result in colored versus colorless seeds, wx+ and wx in nonwaxy versus waxy endosperm, and sh+ and sh in plump versus shrunken endosperm. When plants grown from seeds heterozygous for each of these pairs of alleles (from crosses of pure breeding colorless waxy plump parents with pure breeding colored, nonwaxy, shrunken parents) were testcrossed with plants from colorless, waxy, shrunken seeds, the progeny seeds were as follows: colorless, nonwaxy, shrunken 55 colorless, nonwaxy, plump 105 colorless, waxy, shrunken 5 colorless, waxy, plump 334 colored, waxy, shrunken 98 colored, waxy, plump 61 colored, nonwaxy, shrunken 339 colored, nonwaxy, plump 3 Total: 1000 a. What is the order of the 3 genes? c, sh, wx b. What is the distance in map units between genes c and sh? 55+61+5+3 = 124/1000 x 100 = 12.4 mu c. What is the distance in map units between genes wx and c? 55+61+10+6+105+98 = 335/1000 x 100 = 33.5 mu d. What is the distance in map units between genes wx and sh? 105+98+5+3=211/1000 x 100 = 21.1 mu e. Draw a map to illustrate the distances between the three loci. 33.5 12.4 21.1 c sh wx f. Calculate interference. E= (0.124 x 0.211) x 1000 = 26 C = O/E = 8/26 = 0.308 I = 1-C = 1 – 0.308 = 0.692 or 69.2% positive interference 2 Robinson – Bio 311 Practice Exam 2 3. For each of the following mutations, is it a transition, transversion, addition or deletion? Original DNA Sequence: 5’-GACTACTGTTAA-3’ a. 5’-GACGACTGTTAA-3’ TG transversion pyr pur b. 5’-GACTACTATTAA-3’ GA transition pu pu 4. For each of the following mutations, is it a silent, missense, nonsense or frameshift mutation? Original DNA strand (coding strand) is: 5’-ATGTGGTCTAGAACC-3’ (First amino acid is Met). Met, Trp, Ser, Arg, Thr a. 5’-ATGTGGTCCAGAACC-3’ Silent mutation – UCU & UCC both code for Ser b. 5’-ATGTGGTCTAGAGCC-3’ Missense mutation Thr Ala c. 5’-ATGTGATCTAGAACC-3’ UGA = Stop codon, nonsense mutation 5. During mismatch repair, why is it necessary to distinguish between the template strand and the newly made daughter strand? How is this accomplished? DNA in E.coli is methylated. To distinguish the old template from the newly synthesized strand the mismatch repair mechanism takes advantage of a delay in methylation of the new strand. This makes sense as replication errors produce mismatches only on the newly synthesized strand, so the mismatch repair system replaced the wrong base on that strand. 3 Robinson – Bio 311 Practice Exam 2 6. As you remember, the occurrence of calico cats is due to X-inactivation. a. With this in mind, why is the occurrence of male calico cats unusual? X-inactivation occurs when more than 1 X chromosome is present. Only 1 X chromosome is left active in each cell. Since males are normally XY, there is no X inactivation and thus no calico pattern. b. What kind of aberrant events could give rise to a male calico cat? Describe/draw how such an aberrant event could occur. Nondisjunction at MI in female. XX + Y = XXY X X 7. Five different Hfr strains were allowed to conjugate with F-minus cells. After appropriate screening, it was determined that genes transferred in the following order for each Hfr strain: Strain 1: L N O H I X Strain 2: X I H O Strain 3: X J K A P Strain 4: A P L N O Strain 5: J X I H O N a. What is the order of the genes on the circular bacterial chromosome? L N O H I X J K A P - circular L P N 1 O A 4 5 K 2 H 3 J X I b. For each Hfr strain, give the location of the F factor in the chromosome and its polarity. 4 Robinson – Bio 311 Practice Exam 2 8. A cross is made between 2 E.coli strains: Hfr: met+ bio+ pro+ x F-: met- bio- pro- Interrupted mating studies show that met enters last, so met+ recombinants are selected on medium containing only bio and pro. These recombinants are tested for the presence of bio+ pro+. The following numbers of individuals are found for each genotype: met+ bio+ pro+ met+ bio+ promet+ bio- pro+ met+ bio- pro- 600 2 32 66 a. What is the order of these genes? bio, pro, met b. What is the map distance between pro and bio? (32 + 2)/700 x 100 = 4.86 mu c. What is the map distance between met and pro? (66 + 2)/700 x 100 = 9.71 mu 9. a. Describe and/or illustrate the epigenetic regulation of the Igf2 and H19 locus in a wild-type mouse. Indicate paternal and maternal chromosomes in your discussion/drawing. Maternal chromosome: Insulator able to bind to DMR region, Igf2 gene expression is turned off. H19 gene expression is on. Paternal chromosome: Insulator protein unable to bind DMR due to methylation of the region. The H19 promoter is also disrupted by methylation. Igf2 gene is on, H19 gene is off. 5 Robinson – Bio 311 Practice Exam 2 b. Predict the phenotype of a mouse embryo that is homozygous for a deletion in the DMR (differentially methylated region) of the Igf2 locus. Discuss the affects this deletion would have on the expression of Igf2 and H19. You may assume the deletion of the DMR does not disrupt the promoter of H19. Maternal chromosome: without DMR region insulator cannot bind, both Igf2 and H19 are on. Paternal chromosome: No DMR methylation, but no insulator binding either. Igf2 and H19 are both on. Phenotype of mouse: larger than normal. 10. Consider a heterozygous female fly that contains an inversion spanning three Xlinked genes: forked (f), vermillion (v) and yellow (y). The normal X chromosome is f, v, y and the inversion chromsome is y+, v+, f+. The centromere is not located within the inversion. Note: the lack of + means that each allele is a recessive null mutation and + indicates a normal, wild-type allele. This female is mated to a male that contains the normal X chromosome (no inversion) with all three recessive mutations (f, v, y). Almost all the F1 male progeny are either f, v, y or y+, v+, f+. There are very few recombinants (f, v+, y+ or f, v, y+, etc). Provide an explanation for this suppression of recombination. A diagram might help. Only viable male progeny are of the parental genotype because crossover products contain dicentric and acentric products which are not viable. This is a result of heterozygote paracentric inversion 11. A chromosome has the following segments, where * represents the centromere: ABCDEF*GHI What types of chromosome mutations are required to change this chromosome into each of the following chromosomes? In some cases more than one chromosome mutation may be required. a. A E D C B F * G H IParacentric inversion of BCDE b. A B C D E F * IDeletion of GH c. A B C D E F D E F * G H ITandem duplication DEF d. A B C D E I H G * F Pericentric inversion F*GHI e. A B C C B A D E F * G H I Duplication & paracentric inversion ABC 12. The following diagrams represent two non-homologous chromosomes: ABC*DEFGHI 6 Robinson – Bio 311 Practice Exam 2 TU*VWXYZ What type of chromosome mutation would produce the following chromosomes? 4 pts a. A B C * D E F TU*VWXYZGHI b. Simple translocation ABC*DEFXYZ TU*VWGHI Reciprocal translocation 13. A double-stranded DNA molecule with the sequence shown below produces, in vivo, a polypeptide that is five amino acids long. a. Which strand of DNA is transcribed and in which direction? top strand, right to left b. Label the 5′ and the 3′ ends of each strand. 3’ 5’ 5’ 3’ c. If an inversion occurs between the second and third triplets from the left and right ends, respectively, and the same strand of DNA is transcribed, how long will the resultant polypeptide be? inversion Template strand: 3’-ATG TAC ATC ATT TCA CGG AAT TTC TAG GTA CAT-5’ Coding strand: 5’-TAC ATG TAG TAA AGT GCC TTA AAG ATC CAT GTA-3’ Protein: Met, Stop 1 amino acid long 7