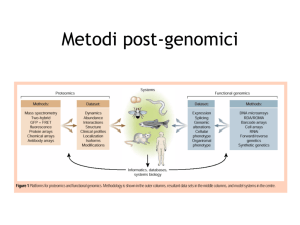
Original Article (Supplementary Information)
advertisement

Original Article (Supplementary Information) Quantitative Proteomic Analysis of Meningiomas for the Identification of Surrogate Protein Markers Samridhi Sharma1†, Sandipan Ray 1†, Aliasgar Moiyadi 2, Epari Sridhar 3 and Sanjeeva Srivastava1* 1 Wadhwani Research Center for Biosciences and Bioengineering, Department of Biosciences and Bioengineering, Indian Institute of Technology Bombay, Powai, Mumbai 400076, India 2 Department of Neurosurgery, Advanced Center for Treatment Research and Education in Cancer, Tata Memorial Center, Kharghar, Navi Mumbai 410210, India 3 Department of Pathology, Tata Memorial Hospital, Mumbai 400012, India. † Both authors contributed equally to the preparation of this manuscript * Correspondence: Dr. Sanjeeva Srivastava, Department of Biosciences and Bioengineering, IIT Bombay, Mumbai 400 076, India: E-mail: sanjeeva@iitb.ac.in Phone: +91-22-2576-7779, Fax: +91-22-2572-3480 Running Title: Proteomic Analysis of Different Grades of Meningioma 1 A 2 3 B 4 5 Figure S1. (A) Representative 2D- DIGE image (Cy3 and Cy5 channel overlap) and trends of differentially expressed proteins in meningioma grade I (compared to healthy controls) visualized in 2DDIGE. (B) Representative 2D- DIGE image (Cy3 and Cy5 channel overlap) Trends of differentially expressed proteins in meningioma grade II (compared to healthy controls) visualized in 2D-DIGE. 6 157 proteins 157 proteins 157 proteins Figure S2. The distribution of the differentially expressed proteins in different grades of meningiomas; grade I (A), II (B) and III (C) identified in iTRAQ. 7 B. Complement pathways 8 B. Complement pathways 9 B. Complement pathways Continued… 10 C. PPAR Signaling Pathway Continued… 11 D. ECM signalling Figure S3. Different biological pathways modulated in meningiomas obtained in DAVID analysis. 12 A 13 B Figure S4. Cellular and molecular sub-trees associated with the differentially expressed proteins identified in meningiomas. 14 Figure S5. Equal loading of protein samples during western blot experiment. Representative CBB stained SDS-PAGE gel (A) and Ponceau stained blot (B) containing the resolved proteins depicting equal loading (50 µg) of the samples (meningioma patients and healthy subjects) in every lane during western blot experiment. 15 Figure S6. Frequency distribution of serum concentrations of Apo E, HPX, Apo A1 and RBP4 in healthy controls and different grades of meningioma patients measured by ELISA. 16 Figure S7. Receiver operating characteristic (ROC) curves depicting accuracy of 4 classifier proteins; Apo E, HPX, Apo A1 and RBP4 for prediction of grade I and grade II meningiomas 17 Figure S8. Comparison of the fold changes of the differentially expressed proteins identified from iTRAQ data with proteins reported in published literature in tissue and CSF samples. Differential expression of proteins in tissue and CSF samples are obtained from reference number 17 and 20 respectively. 18 Supplementary Tables Table S1. Demographics and clinical details of the meningioma patients and healthy subjects Sl.No. Patient ID Diagnosis Grade Age Sex 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 CJ 14742 CJ4231 CH 30545 CF 4450 CH 24953 CJ 20717 CJ 20619 CJ 29822 CJ 29452 CJ 29583 CJ 9179 CJ 26538 CK 726 CK 985 CJ 15491 CJ 15753 CH 17967 CJ 3577 CK 7710 CJ6540 Meningothelial Meningioma Transitional Meningioma Transitional meningioma MeningothelialMeningioma Transitional meningioma MeningothelialMeningioma MeningothelialMeningioma Transitional meningioma Transitional meningioma Meningothelial meningioma Transitional meningioma Lipomatous meningioma MeningothelialMeningioma Angiomatous meningioma Atypical meningioma Atypical meningiom Atypical meningioma Atypical meningioma Atypical meningioma Pappalary Meningioma Gr. I Gr. I Gr. I Gr. I Gr. I Gr. I Gr. I Gr. I Gr. I Gr. I Gr. I Gr. I Gr. I Gr. I Gr. II Gr. II Gr. II Gr. II Gr. II Gr. III 29 35 17 55 41 39 41 50 42 44 57 45 56 55 67 46 58 43 55 61 M F M F F F F F F F F F F F M F M F F M 19 Table S2. Details of all statistically significant (p < 0.05) differentially expressed protein spots in meningioma grade I (compared to healthy controls) visualized in 2D-DIGE Sl. No. Master No. Appearance T-test Av. Ratio 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 684 441 263 277 262 264 707 1312 653 677 758 1009 261 214 657 436 1054 58 1011 1553 1016 322 266 56 510 210 43 1595 590 319 177 1394 36 459 48 59 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 6 (9) 6 (9) 6 (9) 6 (9) 6 (9) 6 (9) 6 (9) 6 (9) 6 (9) 6 (9) 6 (9) 6 (9) 6 (9) 6 (9) 6 (9) 0.00039 0.00089 0.0011 0.0011 0.0013 0.0019 0.0036 0.0054 0.0084 0.012 0.012 0.012 0.013 0.019 0.019 0.02 0.021 0.028 0.035 0.00042 0.0011 0.0015 0.0045 0.0057 0.0075 0.018 0.019 0.021 0.022 0.027 0.032 0.032 0.034 0.034 0.037 0.037 -6.23 4.42 -3.88 -4.04 -3.76 -3.4 2.98 2.84 2.42 -2.67 2.37 1.63 -2.02 1.86 -2.7 2.08 1.6 1.56 1.76 5.49 3.2 -1.4 -2.55 -2.31 -6.23 -3.95 -1.62 6.57 3.08 -1.63 -2.93 2.52 -4.13 1.56 -3.46 -2.73 20 37 38 39 40 41 81 38 73 970 258 6 (9) 6 (9) 6 (9) 6 (9) 6 (9) 21 0.037 0.043 0.044 0.046 0.047 -3.99 3.96 4.17 2.43 2.99 Table S3. Details of all statistically significant (p < 0.05) differentially expressed protein spots in meningioma grade II (compared to healthy controls) visualized in 2D-DIGE Sl. No. Master No. Appearance T-test Av. Ratio 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 1084 632 1340 1483 1435 636 612 641 861 945 1194 682 928 724 915 697 492 901 582 251 651 993 921 587 9 (9) 6 (9) 9 (9) 9 (9) 9 (9) 9 (9) 9 (9) 6 (9) 9 (9) 9 (9) 9 (9) 6 (9) 9 (9) 9 (9) 9 (9) 6 (9) 6 (9) 9 (9) 6 (9) 6 (9) 9 (9) 6 (9) 9 (9) 9 (9) 0.00067 0.0039 0.0071 0.0083 0.011 0.012 0.016 0.019 0.019 0.024 0.024 0.025 0.027 0.029 0.029 0.031 0.033 0.035 0.037 0.042 0.042 0.042 0.049 0.05 1.74 1.49 6.19 -2.01 -2.23 2.06 -3.16 -1.50 3.83 -2.62 -1.9 1.82 1.55 2.83 1.86 1.65 -1.71 1.53 1.47 2.09 -2.62 -1.52 1.62 -2.02 22 Table S4. Master tables for all MALDI-TOF/TOF identified proteins in meningioma grade I visualized in 2D-DIGE# [Analysis Type: Combined (MS+MS/MS); Database: SwissProt; Taxonomy: Human] Sl No. Mast er ID Foldchange Name of protein with UniProt accession number MW (kDa) Protein score No. of matched peptides 1 210 -3.95 (P02768) Serum albumin 69.32 456 30 2 459 1.56 (P00738) Haptoglobin 45.17 217 13 3 1016 3.2 (P02647) Apolipoprotein AI (Apo-AI) 30.75 486 20 23 Peptide sequence CCKADDK YLYEIAR LCTVATLR DDNPNLPR LKCASLQK FQNALLVR LVNEVTEFAK FKDLGEENFK HPDYSVVLLLR AVMDDFAAFVEK RHPDYSVVLLLR CCAAADPHECYAK DVFLGMFLYEYAR KVPQVSTPTLVEVSR QEPERNECFLQHK HPYFYAPELLFFAK RHPYFYAPELLFFAK HPYFYAPELLFFAKR RPCFSALEVDETYVPK VFDEFKPLVEEPQNLIK EFNAETFTFHADICTLSEK ALVLIAFAQYLQQCPFEDHVK TCVADESAENCDKSLHTLFGDK MPCAEDYLSVVLNQLCVLHEK EFNAETFTFHADICTLSEKER QNCELFEQLGEYKFQNALLVR RMPCAEDYLSVVLNQLCVLHEK LVRPEVDVMCTAFHDNEETFLK RMPCAEDYLSVVLNQLCVLHEK LVRPEVDVMCTAFHDNEETFLKK DYAEVGR QLVEIEK GSFPWQAK ILGGHLDAK VGYVSGWGR VMPICLPSK VTSIQDWVQK DIAPTLTLYVGK SCAVAEYGVYVK YVMLPVADQDQCIR VSVNERVMPICLPSK VMPICLPSKDYAEVGR SPVGVQPILNEHTFCAGMSK QKVEPLR AELQEGAR LHELQEK AKPALEDLR LSPLGEEMR QKLHELQEK 4 1011 1.76 (P02753) Plasma retinolbinding protein (RBP4) 23.02 637 11 5 262 -3.76 (P04217) Alpha-1Bglycoprotein precursor (Alpha-1-BN glycoprotein) 54.3 900 16 6 1312 2.84 (P02790) Hemopexin precursor (Beta-1Bglycoprotein) 51.64 239 14 7 1054 1.6 (P02743) Serum amyloid Pcomponent precursor (SAP) 25.37 179 7 24 QGLLPVLESFK DLATVYVDVLK VQPYLDDFQK WQEEMELYR THLAPYSDELR LSPLGEEMRDR VQPYLDDFQKK VSFLSALEEYTK DYVSQFEGSALGK KWQEEMELYR VEPLRAELQEGAR LLDNWDSVTSTFSK DSGRDYVSQFEGSALGK LREQLGPVTQEFWDNLEK QEELCLAR FSGTWYAMAK DPNGLPPEAQK YWGVASFLQK QRQEELCLAR LIVHNGYCDGR LLNLDGTCADSYSFVFSR LLNNWDVCADMVGTFTDTEDPAK GNDDHWIVDTDYDTYAVQYSCR *DPEGLFLQDNIVAEFSVDETGQMSATA K* *KDPEGLFLQDNIVAEFSVDETGQMSAT AK* GVTFLLR LLELTGPK CLAPLEGAR ATWSGAVLAGR LETPDFQLFK SGLSTGWTQLSK HQFLLTGDTQGR CEGPIPDVTFELLR NGVAQEPVHLDSPAIK LELHVDGPPPRPQLR VTLTCVAPLSGVDFQLR IFFHLNAVALGDGGHYTCR SLPAPWLSMAPVSWITPGLK TPGAAANLELIFVGPQHAGNYR SWVPHTFESELSDPVELLVAES *LHDNQNGWSGDSAPVELILSDETLPAP EFSPEPESGR* LHIMAGR LWWLDLK RLWWLDLK DYFMPCPGR VWVYPPEKK FDPVRGEVPPR YYCFQGNQFLR GECQAEGVLFFQGDR ALPQPQNVTSLLGCTH LYLVQGTQVYVFLTK SGAQATWTELPWPHEK LLQDEFPGIPSPLDAAVECHR EVGTPHGIILDSVDAAFICPGSSR *SLGPNSCSANGPGLYLIHGPNLYCYSD VEK* AYSDLSR VGEYSLYIGR QGYFVEAQPK GYVIIKPLVWV IVLGQEQDSYGGK AYSLFSYNTQGR IVLGQEQDSYGGKFDR 8 758 2.37 (P01023) Alpha-2macroglobulin 163.17 318 39 9 707 2.98 (P00450) Ceruloplasmin 122.13 460 36 25 YGAATFTR FQVDNNNR QGIPFFGQVR LPPNVVEESAR YDVENCLANK AIGYLNTGYQR VGFYESDVMGR VTAAPQSVCALR NALFCLESAWK HYDGSYSTFGER NQGNTWLTAFVLK LVHVEEPHTETVR ALLAYAFALAGNQDK IAQWQSFQLEGGLK DNSVHWERPQKPK TEHPFTVEEFVLPK SSSNEEVMFLTVQVK ALLAYAFALAGNQDKR AHTSFQISLSVSYTGSR LLIYAVLPTGDVIGDSAK QFSFPLSSEPFQGSYK VSVQLEASPAFLAVPVEK GHFSISIPVKSDIAPVAR FSGQLNSHGCFYQQVK GNRIAQWQSFQLEGGLK AFQPFFVELTMPYSVIR LLLQQVSLPELPGEYSMK VDLSFSPSQSLPASHAHLR AFQPFFVELTMPYSVIR MCPQLQQYEMHGPEGLR AGAFCLSEDAGLGISSTASLR MCPQLQQYEMHGPEGLR LHTEAQIQEEGTVVELTGR QQNAQGGFSSTQDTVVALHALSK SLFTDLEAENDVLHCVAFAVPK NALFCLESAWKTAQEGDHGSHVYTK VVSMDENFHPLNELIPLVYIQDPK *YDVENCLANKVDLSFSPSQSLPASHAH LR* *SPCYGYQWVSEEHEEAHHTAYLVFSPS K* KGSLHANGR QYTDSTFR EYTDASFTNR EYTDASFTNRK GAYPLSIEPIGVR EVGPTNADPVCLAK QSEDSTFYLGER DIASGLIGPLIICK ALYLQYTDETFR VNKDDEEFIESNK DLYSGLIGPLIVCR RQSEDSTFYLGER QYTDSTFRVPVER KALYLQYTDETFR NNEGTYYSPNYNPQSR TTIEKPVWLGFLGPIIK TYYIAAVEVEWDYSPQR NLASRPYTFHSHGITYYK LISVDTEHSNIYLQNGPDR MHSMNGFMYGNQPGLTMCK KAEEEHLGILGPQLHADVGDK FNKNNEGTYYSPNYNPQSR KLISVDTEHSNIYLQNGPDR MYYSAVDPTKDIFTGLIGPMK HYYIGIIETTWDYASDHGEK MYSVNGYTFGSLPGLSMCAEDR MFTTAPDQVDKEDEDFQESNK GPEEEHLGILGPVIWAEVGDTIR SGAGTEDSACIPWAYYSTVDQVK HYYIGIIETTWDYASDHGEKK SVPPSASHVAPTETFTYEWTVPK GVYSSDVFDIFPGTYQTLEMFPR ERGPEEEHLGILGPVIWAEVGDTIR WYLFGMGNEVDVHAAFFHGQALTNK *ADDKVYPGEQYTYMLLATEEQSPGEG DGNCVTR* *NMATRPYSIHAHGVQTESSTVTPTLPG ETLTYVWK* 10 214 1.86 (P10909) Clusterin precursor (Complementassociated protein SP-40) 52.46 151 8 11 1553 5.49 (P02750) Leucine-rich alpha-2-glycoprotein (LRG) 38.15 637 10 12 970 2.43 (P02790) Hemopexin precursor (Beta-1Bglycoprotein) 52.38 207 9 13 322 -1.4 (P02774) Vitamin D Binding protein 199 3 14 653 2.42 (P01009) Alpha-1antitrypsin 46.87 355 5 15 1394 2.52 (P01024) Complement C3 188.56 275 8 16 258 2.99 (P02649) Apolipoprotein E precursor 36.24 234 6 54.52 26 VCRSGSGLVGR EGDDDRTVCR ELDESLQVAER NPKFMETVAEK ASSIIDELFQDR EILSVDCSTNNPSQAK LFDSDPITVTVPVEVSR VTTVASHTSDSDVPSGVTEVVVK GPLQLER DCQVFR VAAGAFQGLR ALGHLDLSGNR DLLLPQPDLR ENQLEVLEVSWLHGLK TLDLGENQLETLPPDLLR NALTGLPPGLFQASATLDTLVLK DGFDISGNPWICDQNLSDLYR LQELHLSSNGLESLSPEFLRPVPQLR RLWWLDLK DYFMPCPGR DYFM*PCPGR NFPSPVDAAFR FDPVRGEVPPR YYCFQGNQFLR GECQAEGVLFFQGDR LLQDEFPGIPSPLDAAVECHR EVGTPHGIILDSVDAAFICPGSSR SCESNSPFPVHPGTAECCTK EVVSLTEACCAEGADPDCYDTR HQPQEFPTYVEPTNDEICEAFR GKWERPFEVK ITPNLAEFAFSLYR FNKPFVFLMIEQNTK ELDRDTVFALVNYIFFK LYHSEAFTVNFGDTEEAKK GYTQQLAFR DFDFVPPVVR KGYTQQLAFR AGDFLEANYM*NLQR SYTVAIAGYALAQMGR SYTVAIAGYALAQMGR ILLQGTPVAQMTEDAVDAER EGVQKEDIPPADLSDQVPDTESETR LAVYQAGAR LGPLVEQGR LQAEAFQAR AKLEEQAQQIR 17 319 -1.63 (P02768) Serum albumin precursor 69.32 663 26 18 436 2.08 (P00738) Haptoglobin precursor 45.17 248 12 19 441 4.42 (P00738) Haptoglobin precursor 45.17 713 14 20 1009 1.63 (P10909) Clusterin precursor (Complementassociated protein SP-40) 52.46 179 8 21 261 -2.02 (P02768) Serum albumin precursor 69.3 674 23 27 AATVGSLAGQPLQER SELEEQLTPVAEETR CCKADDK YLYEIAR D DNPNLPR FQNALLVR CASLQKFGER HPDYSVVLLLR AVMDDFAAFVEK RHPDYSVVLLLR CCAAADPHECYAK DVFLGMFLYEYAR KVPQVSTPTLVEVSR QNCELFEQLGEYK QEPERNECFLQHK HPYFYAPELLFFAK RPCFSALEVDETYVPK NECFLQHKDDNPNLPR VFDEFKPLVEEPQNLIK VHTECCHGDLLECADDR EFNAETFTFHADICTLSEK MPCAEDYLSVVLNQLCVLHEK EFNAETFTFHADICTLSEKER VHTECCHGDLLECADDRADLAK LVRPEVDVMCTAFHDNEETFLK RMPCAEDYLSVVLNQLCVLHEK LVRPEVDVMCTAFHDNEETFLKK CCTESLVNRRPCFSALEVDETYVPK NYYKLR NPANPVQR VMPICLPSK YQCKNYYK LPECEAVCGKPK TEGDGVYTLNDKK LRTEGDGVYTLNDK YVMLPVADQDQCIR LRTEGDGVYTLNNEK AVGDKLPECEAVCGKPK LPECEADDGCPKPPEIAHGYVEHSVR *AVGDKLPECEADDGCPKPPEIAHGYVE HSVR* DYAEVGR GSFPWQAK VGYVSGWGR VTSIQDWVQK DIAPTLTLYVGK TEGDGVYTLNDK SCAVAEYGVYVK YVMLPVADQDQCIR VSVNERVMPICLPSK VVLHPNYSQVDIGLIK VMPICLPSKDYAEVGR SPVGVQPILNEHTFCAGMSK YVMLPVADQDQCIRHYEGSTVPEK *YQEDTCYGDAGSAFAVHDLEEDTWY ATGILSFDK* IDSLLENDR RPHFFFPK QTCMKFYAR TLLSNLEEAKK NPKFMETVAEK ASSIIDELFQDR QQTHMLDVMQDHFSR EPQDTYHYLPFSLPHR YLYEIAR FQNALLVR QTALVELVK CASLQKFGER ECCEKPLLEK HPDYSVVLLLR AVMDDFAAFVEK RHPDYSVVLLLR CCAAADPHECYAK VPQVSTPTLVEVSR DVFLGMFLYEYAR YICENQDSISSKLK KVPQVSTPTLVEVSR QNCELFEQLGEYK QEPERNECFLQHK RPCFSALEVDETYVPK VPQVSTPTLVEVSRNLGK SLHTLFGDKLCTVATLR AAFTECCQAADKAACLLPK VFDEFKPLVEEPQNLIK EFNAETFTFHADICTLSEK MPCAEDYLSVVLNQLCVLHEK RMPCAEDYLSVVLNQLCVLHEK # Alterations in protein expression levels in MG I patients were measured using healthy subjects as controls * Indicates the continuation of peptide sequence in the next line 28 Table S5. Master tables for all MALDI-TOF/TOF identified proteins in meningioma grade II visualized in 2D-DIGE # [Analysis Type: Combined (MS+MS/MS); Database: SwissProt; Taxonomy: Human] Sl No. Mast er ID Foldchange Name of protein with UniProt accession number MW (kDa) Protein score No. of matched peptide s Peptide sequence 1 636 2.06 (P00738) Haptoglobin 45.17 713 14 2 641 -1.50 (P00751) Complement factor B precursor (EC 3.4.21.47) 85.4 717 26 3 651 -2.62 (P02787) Serotransferrin precursor (Transferrin) 76.99 1160 42 DYAEVGR GSFPWQAK VGYVSGWGR VTSIQDWVQK DIAPTLTLYVGK TEGDGVYTLNDK SCAVAEYGVYVK YVMLPVADQDQCIR VSVNERVMPICLPSK VVLHPNYSQVDIGLIK VMPICLPSKDYAEVR SPVGVQPILNEHTFCAGMSK YVMLPVADQDQCIRHYEGSTVPK *YQEDTCYGDAGSAFAVHDLEEDTWYA TGILSFDK* ISVIRPSK STGSWSTLK VASYGVKPR DISEVVTPR EELLPAQDIK ALFVSEEEKK VKDISEVVTPR YGLVTYATYPK LPPTTTCQQQK EKLQDEDLGFL LEDSVTYHCSR QLNEINYEDHK DFHINLFQVLPWLK FIQVGVISWGVVDVCK EAGIPEFYDYDVALIK FLCTGGVSPYADPNTCR KEAGIPEFYDYDVALIK SRFIQVGVISWGVVDVK DLEIEVVLFHPNYNINGK YGQTIRPICLPCTEGTTR AIHCPRPHDFENGEYWPR LPPTTTCQQQKEELLPAQDIK WSGQTAICDNGAGYCSNPGIPIGTR LLQEGQALEYVCPSGFYPYPVQTR EDYLDVYVFGVGPLVNQVNINALASK *GHESCMGAVVSEYFVLTAAHCFTVD D KEHSIK* NPDPWAK SCHTAVGR APNHAVVTR ASYLDCIR YLGEEYVK KASYLDCIR WCALSHHER DSGFQMNQLR SASDLTWDNK HSTIFENLANK EFQLFSSPHGK EGYYGYTGAR WCAVSEHEAT K 29 4 1084 1.74 (P00738) Haptoglobin 45.17 306 9 5 697 1.65 (P02790) Hemopexin (Beta1B-glycoprotein) 51.64 596 21 6 682 1.82 (P01876) Ig alpha-1 chain C region 37.63 491 14 30 KDSGFQMNQLR DYELLCLDGTR KSASDLTWDNLK SVIPSDGPSVACVK MYLGYEYVTAIR SKEFQLFSSPHGK LKCDEWSVNSVGK CSTSSLLEACTFR DQYELLCLDNTR DLLFRDDTVCLAK FDEFFSEGCAPGSK TAGWNIPMGLLYNK KPVEEYANCHLAR EDPQTFYYAVAVVK KCSTSSLLEACTFR DCHLAQVPSHTVVAR FDEFFSEGCAPGSKK IECVSAETTEDCIAK EGTCPEAPTDECKPVK WCAVSEHEATKCQSFR ADRDQYELLCLDNTR NLNEKDYELLCLDGTR EDLIWELLNQAQEHFGK SDNCEDTPEAGYFAVAVVK SAGWNIPIGLLYCDLPEPR NLREGTCPEAPTDECKPVK KPVDEYKDCHLAQVPSHTVVAR LCMGSGLNLCEPNNKEGYYGYTGAFR *AIAANEADAVTLDAGLVYDAYLAPNN LKPVVAEFYGSK* DYAEVGR NYYKLR GSFPWQAK VGYVSGWGR VMPICLPSK YQCKNYYK VTSIQDWVQK SCAVAEYGVYVK YVMLPVADQDQCIR LHIMAGR LWWLDLK GEVPPRYPR RLWWLDLK DYFMPCPGR VWVYPPEKK FDPVRGEVPPR GDKVWVYPPEK SWPAVGNCSSALR YYCFQGNQFLR EWFWDLATGTMK GECQAEGVLFFQGDR LYLVQGTQVYVFLTK SGAQATWTELPWPHEK SWPAVGNCSSALRWLGR LLQDEFPGIPSPLDAAVECHR EVGTPHGIILDSVDAAFICPGSSR CSPHLVLSALTSDNHGATYAFSGTHYWR GECQAEGVLFFQGDREWFWDLATGTM K *DGWHSWPIAHQWPQGPSAVDAAFSWE EK* *SLGPNSCSANGPGLYLIHGPNLYCYSDV EK* VAAEDWK YLTWASR SAVQGPPER EKYLTWASR WLQGSQELPR TFTCTAAYPESK 7 612 -3.16 (P02768) Serum albumin 69.3 1410 26 8 861 3.83 (P02647) Apolipoprotein AI (Apo-AI) 30.75 486 20 9 587 -2.02 (P02768) Serum albumin 69.32 456 30 31 DASGVTFTWTPSSGK QEPSQGTTTFAVTSILR GDTFSCMVGHEALPLAFTQK TFTCTAAYPESKTPLTATLSK KGDTFSCMVGHEALPLAFTQK DLCGCYSVSSVLPGCAEPWNHGK *NFPPSQDASGDLYTTSSQLTLPATQCLA GK* *SGNTFRPEVHLLPPPSEELALNELVTLT CLAR* YLYEIAR LCTVATLR DDNPNLPR FQNALLVR NECFLQHK CCTESLVNR LVNEVTEFAK ECCEKPLLEK HPDYSVVLLLR AVMDDFAAFVEK AAFTECCQAADK RHPDYSVVLLLR CCAAADPHECYAK VPQVSTPTLVEVSR DVFLGMFLYEYAR KVPQVSTPTLVEVSR QNCELFEQLGEYK HPYFYAPELLFFAK RPCFSALEVDETYVPK VHTECCHGDLLECADDR VFDEFKPLVEEPQNLIK EFNAETFTFHADICTLSEK ALVLIAFAQYLQQCPFEDHVK MPCAEDYLSVVLNQLCVLHEK LVRPEVDVMCTAFHDNEETFLK SHCIAEVENDEMPADLPSLAADFVESK QKVEPLR AELQEGAR LHELQEK AKPALEDLR LSPLGEEMR QKLHELQEK QGLLPVLESFK DLATVYVDVLK VQPYLDDFQK WQEEMELYR THLAPYSDELR LSPLGEEMRDR VQPYLDDFQKK VSFLSALEEYTK DYVSQFEGSALGK KWQEEMELYR VEPLRAELQEGAR LLDNWDSVTSTFSK DSGRDYVSQFEGSALGK LREQLGPVTQEFWDNLEK CCKADDK YLYEIAR LCTVATLR DDNPNLPR LKCASLQK FQNALLVR LVNEVTEFAK FKDLGEENFK HPDYSVVLLLR AVMDDFAAFVEK RHPDYSVVLLLR CCAAADPHECYAK DVFLGMFLYEYAR 10 945 -2.62 (P02765) Alpha-2-HSglycoprotein 39.29 495 10 KVPQVSTPTLVEVSR QEPERNECFLQHK HPYFYAPELLFFAK RHPYFYAPELLFFAK HPYFYAPELLFFAKR RPCFSALEVDETYVPK VFDEFKPLVEEPQNLIK EFNAETFTFHADICTLSEK ALVLIAFAQYLQQCPFEDHVK TCVADESAENCDKSLHTLFGDK MPCAEDYLSVVLNQLCVLHEK EFNAETFTFHADICTLSEKER QNCELFEQLGEYKFQNALLVR RMPCAEDYLSVVLNQLCVLHEK LVRPEVDVMCTAFHDNEETFLK RMPCAEDYLSVVLNQLCVLHEK LVRPEVDVMCTAFHDNEETFLKK QYGFCK FSVVYAK HTLNQIDEVK CDSSPDSAEDVRK EHAVEGDCDFQLLK HTFMGVVSLGSPSGEVSHPR HTFMGVVSLGSPSGEVSHPRK AQLVPLPPSTYVEFTVSGTDCVAK *QPNCDDPETEEAALVAIDYINQNLPWG YK* *VWPQQPSGELFEIEIDTLETTCHVLDPTP VAR* # Alterations in protein expression levels in MG II patients were measured using healthy subjects as controls * Indicates the continuation of peptide sequence in the next line 32 Table S6. Complete details of protein identification and quantitative iTRAQ data for different grades of meningiomas. Gr ou p N u m Nu m Sp ect ra Nu m Pe ps Un iq ue Sc or e Un iq ue % Cov era ge log2_iTRAQ _115_114_m edian Fo ldch an ge (H C vs. M G1 ) log2_iTRAQ _116_114_m edian Fo ldch an ge (H C vs. M G2 ) log2_iTRAQ _117_114_m edian Fo ldch an ge (H C vs. M G3 ) Accessi on_num ber Protein_n ame Peptide sequence 1 320 100 179 7.2 8 32.7 0.843 1.7 9 1.895 3.7 2 2.718 6.5 8 P04114. 2 Apolipopr otein B100 AALTELSLGSAYQA MILGVDSK ADSVVDLLSYNVQG SGETTYDHK AEPLAFTFSHDYK AHLDIAGSLEGHLR ALVEQGFTVPEIK AQNLYQELLTQEGQ ASFQGLK ATFQTPDFIVPLTDLR ATGVLYDYVNK ATVAVYLESLQDTK AVSMPSFSILGSDVR DAVEKPQEFTIVAFV K DFSLWEK DKAQNLYQELLTQE GQASFQGLK DKDQEVLLQTFLDD ASPGDKR DLKVEDIPLAR DNVFDGLVR EELCTMFIR EFNLQNMGLPDFHIP ENLFLK EFQVPTFTIPK ESQLPTVMDFR EVGTVLSQVYSK EVYGFNPEGK EYSGTIASEANTYLN SK FDHTNSLNIAGLSLD FSSK FPEVDVLTK FSDEGTHESQISFTIE GPLTSFGLSNK GFEPTLEALFGK GIISALLVPPETEEAK GMALFGEGK HSITNPLAVLCEFISQ SIK IADFELPTIIVPEQTIEI PSIK IAELSATAQEIIK IDDIWNLEVK IEFEWNTGTNVDTK IEGNLIFDPNNYLPK IEIPLPFGGK IGQDGISTSATTNLK ILGEELGFASLHDLQ LLGK INCKVELEVPQLCSFI LK ITENDIQIALDDAK ITLPDFR IVQILPWEQNEQVK KMTSNFPVDLSDYPK 33 KYTYNYEAESSSGVP GTADSR LDFSSQADLR LEIQSQVDSQHVGHS VLTAK LELELRPTGEIEQYSV SATYELQR LELELRPTGEIEQYSV SATYELQREDR LIDVISMYR LLLQMDSSATAYGST VSK LNDLNSVLVMPTFH VPFTDLQVPSCK LNTDIAGLASAIDMS TNYNSDSLHFSNVFR LPQQANDYLNSFNW ER LPYTIITTPPLKDFSL WEK LSLESLTSYFSIESSTK LSLESLTSYFSIESSTK GDVK LSLPDFK LSNDMMGSYAEMK LTLDIQNK LVGFIDDAVK MGLAFESTK MTSNFPVDLSDYPK MYQMDIQQELQR NFVASHIANILNSEEL DIQDLK NFVASHIANILNSEEL DIQDLKK NIQEYLSILTDPDGK NLQDLLQFIFQLIEDN IK NLQNNAEWVYQGAI R NLTDFAEQYSIQDWA K NSLFFSAQPFEITAST NNEGNLK QTIIVVLENVQR QVFLYPEKDEPTYIL NIKR SGSSTASWIQNVDTK SLWDFLK SVSDGIAALDLNAVA NK SVSLPSLDPASAK TEHGSEMLFFGNAIE GK TEVIPPLIENR TFQIPGYTVPVVNVE VSPFTIEMSAFGYVFP K TIHDLHLFIENIDFNK TILGTMPAFEVSLQA LQK TLADLTLLDSPIK TLADLTLLDSPIKVPL LLSEPINIIDALEMR TLQGIPQMIGEVIR TSSFALNLPTLPEVK VEDIPLAR VELEVPQLCSFILK VHANPLLIDVVTYLV ALIPEPSAQQLREIFN MAR VIGNMGQTMEQLTP ELK VLLDQLGTTISFER VNWEEEAASGLLTSL K VNWEEEAASGLLTSL 34 KDNVPK VPLLLSEPINIIDALE MR VPQTDMTFR VPSYTLILPSLELPVL HVPR VSALLTPAEQTGTW K YDKNQDVHSINLPFF ETLQEYFER YENYELTLK YSQPEDSLIPFFEITVP ESQLTVSQFTLPK YTYNYEAESSSGVPG TADSR 35 2 393 75 134 4.3 6 54.9 0.316 1.2 4 1.323 2.5 0 36 1.591 3.0 1 P01024. 2 Complem ent C3 ACEPGVDYVYK ACEPGVDYVYKTR ADIGCTPGSGKDYAG VFSDAGLTFTSSSGQ QTAQR AEDLVGKSLYVSAT VILHSGSDMVQAER AGDFLEANYMNLQR APSTWLTAYVVKVF SLAVNLIAIDSQVLC GAVK AYYENSPQQVFSTEF EVK DAPDHQELNLDVSL QLPSR DFDFVPPVVR DICEEQVNSLPGSITK DSCVGSLVVK DSITTWEILAVSMSD K DSITTWEILAVSMSD KK DYAGVFSDAGLTFTS SSGQQTAQR ENEGFTVTAEGK EPGQDLVVLPLSITTD FIPSFR EVVADSVWVDVK EVVADSVWVDVKDS CVGSLVVK EYVLPSFEVIVEPTEK FISLGEACK FVTVQATFGTQVVE K FYYIYNEK GLEVTITAR ILLQGTPVAQMTEDA VDAER IPIEDGSGEVVLSR ISLPESLK KQELSEAEQATR KVEGTAFVIFGIQDG EQR KVFLDCCNYITELR KVLLDGVQNPRAED LVGK LDKACEPGVDYVYK LDKACEPGVDYVYK TR LESEETMVLEAHDA QGDVPVTVTVHDFP GK LESEETMVLEAHDA QGDVPVTVTVHDFP GKK NNNEKDMALTAFVLI SLQEAK NTLIIYLDK NTMILEICTR QDSLSSQNQLGVLPL SWDIPELVNMGQWK QGALELIK QKPDGVFQEDAPVIH QEMIGGLR QLYNVEATSYALLAL LQLKDFDFVPPVVR QVREPGQDLVVLPLS ITTDFIPSFR RIPIEDGSGEVVLSR SDDKVTLEERLDK SEETKENEGFTVTAE GK SEETKENEGFTVTAE GKGQGTLSVVTMYH AK SEFPESWLWNVEDL K SEFPESWLWNVEDL KEPPK SGIPIVTSPYQIHFTK SGQSEDRQPVPGQQ MTLK SGQSEDRQPVPGQQ MTLKIEGDHGAR SLYVSATVILHSGSD MVQAER SNLDEDIIAEENIVSR SSLSVPYVIVPLK SSLSVPYVIVPLKTGL QEVEVK TELRPGETLNVNFLL R TGLQEVEVKAAVYH HFISDGVR TKKQELSEAEQATR TVMVNIENPEGIPVK VELLHNPAFCSLATT K VFLDCCNYITELR VFLDCCNYITELRR VHQYFNVELIQPGAV K VLLDGVQNPR VLLDGVQNPRAEDL VGK VPVAVQGEDTVQSL TQGDGVAK VQLSNDFDEYIMAIE QTIK VQLSNDFDEYImAIE QTIK VQLSNDFDEYIMAIE QTIKSGSDEVQVGQQ R VSHSEDDCLAFK VTIKPAPETEK VVLVAVDK VYAYYNLEESCTR WLILEK YFKPGMPFDLMVFV TNPDGSPAYR YISKYELDK 37 3 317 52 954 .16 47.8 0.54 1.4 5 1.788 3.4 5 38 1.641 3.1 2 P01023. 3 Alpha-2macroglob ulin AFQPFFVELTMPYSVI R AFQPFFVELTmPYSVI R AGAFCLSEDAGLGIS STASLR ALLAYAFALAGNQD K AVDQSVLLMKPDAE LSASSVYNLLPEK AYIFIDEAHITQALIW LSQR DMYSFLEDMGLK DTVIKPLLVEPEGLE K EEFPFALGVQTLPQT CDEPK ETTFNSLLCPSGGEVS EELSLK ETTFNSLLCPSGGEVS EELSLKLPPNVVEES AR FEVQVTVPK GGVEDEVTLSAYITI ALLEIPLTVTHPVVR HNVYINGITYTPVSST NEK HNVYINGITYTPVSST NEKDMYSFLEDMGL K HYDGSYSTFGER IAQWQSFQLEGGLK IAQWQSFQLEGGLKQ FSFPLSSEPFQGSYK KDTVIKPLLVEPEGL EK KYSDASDCHGEDSQ AFCEK LHTEAQIQEEGTVVE LTGR LLIYAVLPTGDVIGDS AK LLLQQVSLPELPGEY SMK LPPNVVEESAR LVHVEEPHTETVR MCPQLQQYEMHGPE GLR NALFCLESAWK NEDSLVFVQTDK QFSFPLSSEPFQGSYK QGIPFFGQVR QQNAQGGFSSTQDT VVALHALSK QTVSWAVTPK SASNMAIVDVK SGGRTEHPFTVEEFV LPK SGGRTEHPFTVEEFV LPKFEVQVTVPK SLFTDLEAENDVLHC VAFAVPK SPCYGYQWVSEEHE EAHHTAYLVFSPSK SSSNEEVMFLTVQVK TEHPFTVEEFVLPK TEVSSNHVLIYLDK TTVMVKNEDSLVFV QTDK VDLSFSPSQSLPASHA HLR VGFYESDVMGR VSVQLEASPAFLAVP VEK VTAAPQSVCALR VTGEGCVYLQTSLK VVSMDENFHPLNELI PLVYIQDPK VVSMDENFHPLNELI PLVYIQDPKGNR VYDYYETDEFAIAEY NAPCSK YDVENCLANK YDVENCLANKVDLS FSPSQSLPASHAHLR YNILPEKEEFPFALGV QTLPQTCDEPK YSDASDCHGEDSQAF CEK 39 4 382 46 887 .4 59.1 -0.573 0.6 7 0.095 1.0 7 40 0.332 1.2 6 P02787. 3 Serotransf errin ADRDQYELLCLDNT R ASYLDCIR CDEWSVNSVGK CLKDGAGDVAFVK CLVEKGDVAFVK CSTSSLLEACTFR DGAGDVAFVK DLLFK DLLFKDSAHGFLK DLLFRDDTVCLAK DSGFQMNQLR EDLIWELLNQAQEHF GK EDLIWELLNQAQEHF GKDK EDPQTFYYAVAVVK EDPQTFYYAVAVVK K EFQLFSSPHGK EGTCPEAPTDECKPV K EGTCPEAPTDECKPV KWCALSHHER EGYYGYTGAFR FDEFFSEGCAPGSK FDEFFSEGCAPGSKK GDVAFVK HSTIFENLANK HSTIFENLANKADRD QYELLCLDNTR IECVSAETTEDCIAK INHCRFDEFFSEGCAP GSK KPVDEYKDCHLAQV PSHTVVAR KPVEEYANCHLAR KSASDLTWDNLK LCMGSGLNLCEPNN K LCMGSGLNLCEPNN KEGYYGYTGAFR LKCDEWSVNSVGK LKCDEWSVNSVGKIE CVSAETTEDCIAK MYLGYEYVTAIR mYLGYEYVTAIR NLNEKDYELLCLDGT R NLNEKDYELLCLDGT RKPVEEYANCHLAR SAGWNIPIGLLYCDL PEPR SASDLTWDNLK SASDLTWDNLKGK SETKDLLFRDDTVCL AK SMGGKEDLIWELLN QAQEHFGK SmGGKEDLIWELLNQ AQEHFGK SMGGKEDLIWELLN QAQEHFGKDK SmGGKEDLIWELLNQ AQEHFGKDK SVIPSDGPSVACVK TAGWNIPMGLLYNK WCAVSEHEATK YLGEEYVK 5 186 42 774 .94 31 0.245 1.1 9 0.67 1.5 9 41 0.727 1.6 6 P0C0L5 .1 Complem ent C4-B AEFQDALEK AEFQDALEKLNMGIT DLQGLR AEMADQASAWLTR DFALLSLQVPLK DFALLSLQVPLKDAK DSSTWLTAFVLK ECVGFEAVQEVPVGL VQPASATLYDYYNPE R ECVGFEAVQEVPVGL VQPASATLYDYYNPE RR EELVYELNPLDHR EMSGSPASGIPVK EPFLSCCQFAESLR FGLLDEDGK FGLLDEDGKK FGLLDEDGKKTFFR GLEEELQFSLGSK GPEVQLVAHSPWLK GSFEFPVGDAVSK ITPGKPYILTVPGHLD EMQLDIQAR KEVYMPSSIFQDDFVI PDISEPGTWK KKEVYMPSSIFQDDF VIPDISEPGTWK LELSVDGAK LHLETDSLALVALGA LDTALYAAGSK LLATLCSAEVCQCAE GK LNMGITDLQGLR LQETSNWLLSQQQA DGSFQDLSPVIHR LTVAAPPSGGPGFLSI ERPDSRPPR SFFPENWLWR STQDTVIALDALSAY WIASHTTEER TEQWSTLPPETK VDFTLSSER VDFTLSSERDFALLSL QVPLKDAK VDVQAGACEGK VEYGFQVK VGDTLNLNLR VLQIEKEGAIHREEL VYELNPLDHR VLSLAQEQVGGSPEK VQQPDCREPFLSCCQ FAESLR VQQPDCREPFLSCCQ FAESLRK VTASDPLDTLGSEGA LSPGGVASLLR YLDKTEQWSTLPPET K YVLPNFEVK YVSHFETEGPHVLLY FDSVPTSR 6 108 30 615 .64 67.4 -0.659 0.6 3 0.075 1.0 5 42 -0.723 0.6 1 P02768. 2 Serum albumin ADDKETCFAEEGKK AEFAEVSKLVTDLTK ALVLIAFAQYLQQCP FEDHVK AVMDDFAAFVEK DVFLGMFLYEYAR EFNAETFTFHADICTL SEK EFNAETFTFHADICTL SEKER KVPQVSTPTLVEVSR LKECCEKPLLEK LVAASQAALGL LVNEVTEFAK LVRPEVDVMCTAFH DNEETFLK LVRPEVDVMCTAFH DNEETFLKK LVTDLTK MPCAEDYLSVVLNQ LCVLHEK MPCAEDYLSVVLNQ LCVLHEKTPVSDR NYAEAKDVFLGMFL YEYAR QNCELFEQLGEYK QTALVELVK RMPCAEDYLSVVLN QLCVLHEK RMPCAEDYLSVVLN QLCVLHEKTPVSDR RPCFSALEVDETYVP K SHCIAEVENDEMPAD LPSLAADFVESK SHCIAEVENDEMPAD LPSLAADFVESKDVC K VFDEFKPLVEEPQNLI K VHTECCHGDLLECA DDR VHTECCHGDLLECA DDRADLAK VPQVSTPTLVEVSR YICENQDSISSK YLYEIAR 7 197 29 539 .32 70.4 1.19 2.2 8 2.825 7.0 9 43 3.729 13. 26 P02647. 1 Apolipopr otein A-I AKPALEDLR AKVQPYLDDFQK ATEHLSTLSEK ATEHLSTLSEKAKPA LEDLR DLATVYVDVLK DLATVYVDVLKDSG RDYVSQFEGSALGK DLEEVK DSGRDYVSQFEGSAL GK DYVSQFEGSALGK EQLGPVTQEFWDNL EK EQLGPVTQEFWDNL EKETEGLR KWQEEMELYR LLDNWDSVTSTFSK LLDNWDSVTSTFSKL R LREQLGPVTQEFWD NLEK LREQLGPVTQEFWD NLEKETEGLR LSPLGEEMR LSPLGEEMRDR QGLLPVLESFK QGLLPVLESFKVSFLS ALEEYTK QKVEPLRAELQEGAR THLAPYSDELR VEPLRAELQEGAR VKDLATVYVDVLK VKDLATVYVDVLKD SGR VQPYLDDFQK VQPYLDDFQKKWQE EMELYR VSFLSALEEYTK WQEEMELYR 8 152 27 564 .51 37.8 0.048 1.0 3 1.028 2.0 4 44 0.52 1.4 3 P00450. 1 Ceruloplas min ADDKVYPGEQYTYM LLATEEQSPGEGDGN CVTR AEEEHLGILGPQLHA DVGDK AGLQAFFQVQECNK ALYLQYTDETFR DIASGLIGPLIICK DIFTGLIGPMK DLYSGLIGPLIVCR ERGPEEEHLGILGPVI WAEVGDTIR EVGPTNADPVCLAK GAYPLSIEPIGVR GPEEEHLGILGPVIW AEVGDTIR GVYSSDVFDIFPGTY QTLEMFPR HYYIAAEEIIWNYAP SGIDIFTK HYYIGIIETTWDYAS DHGEKK KAEEEHLGILGPQLH ADVGDK KAEEEHLGILGPQLH ADVGDKVK KERGPEEEHLGILGP VIWAEVGDTIR MFTTAPDQVDKEDE DFQESNK MYYSAVDPTK MYYSAVDPTKDIFTG LIGPMK NNEGTYYSPNYNPQS R QKDVDKEFYLFPTVF DENESLLLEDNIR SGAGTEDSACIPWAY YSTVDQVK SVPPSASHVAPTETFT YEWTVPK TYCSEPEKVDKDNED FQESNR VNKDDEEFIESNK VYPGEQYTYMLLAT EEQSPGEGDGNCVTR 9 153 27 485 .96 32.8 -0.147 0.9 0 0.2 1.1 5 -0.034 0.9 8 P08603. 4 Complem ent factor H 10 126 23 392 .79 53.1 0.761 1.6 9 2.641 6.2 4 2.874 7.3 3 P01009. 3 Alpha-1antitrypsin 45 AGEQVTYTCATYYK AVYTCNEGYQLLGEI NYR CFEGFGIDGPAIAK CTLKPCDYPDIK DGEKVSVLCQENYLI QEGEEITCKDGR DGWSAQPTCIK DTSCVNPPTVQNAYI VSR EIMENYNIALR EKTKEEYGHSEVVEY YCNPR GDAVCTESGWRPLPS CEEK IDVHLVPDR IVSSAMEPDREYHFG QAVR LGYVTADGETSGSIT CGK LSYTCEGGFR NTEILTGSWSDQTYP EGTQAIYK RPCGHPGDTPFGTFT LTGGNVFEYGVK SCDIPVFMNAR SIDVACHPGYALPK SITCIHGVWTQLPQC VAIDK SSNLIILEEHLK TDCLSLPSFENAIPMG EK TGESVEFVCK TKEEYGHSEVVEYY CNPR VSVLCQENYLIQEGE EITCK VSVLCQENYLIQEGE EITCKDGR WQSIPLCVEK WSSPPQCEGLPCK AVLTIDEK AVLTIDEKGTEAAGA MFLEAIPMSIPPEVK DTEEEDFHVDQVTTV K DTEEEDFHVDQVTTV KVPMMK ELDRDTVFALVNYIF FK FLEDVK GTEAAGAMFLEAIPM SIPPEVK ITPNLAEFAFSLYR IVDLVK IVDLVKELDRDTVFA LVNYIFFK KLYHSEAFTVNFGDT EEAK KLYHSEAFTVNFGDT EEAKK LGMFNIQHCK LQHLENELTHDIITK LSITGTYDLK LVDKFLEDVK LVDKFLEDVKK LYHSEAFTVNFGDTE EAK QINDYVEK SPLFMGK SVLGQLGITK TDTSHHDQDHPTFNK ITPNLAEFAFSLYR WERPFEVK 11 83 22 400 .36 18.2 -0.143 0.9 1 0.507 1.4 2 1.168 2.2 5 P02751. 4 Fibronecti n 12 182 22 389 .03 48.2 -0.28 1.6 7 0.133 2.1 0 -0.212 1.8 7 P00738. 1 Haptoglob in 46 DDKESVPISDTIIPAV PPPTDLR DLEVVAATPTSLLIS WDAPAVTVR DLQFVEVTDVK EESPLLIGQQSTVSDV PR EINLAPDSSSVVVSGL MVATK EYLGAICSCTCFGGQ R GEWTCIAYSQLR GFNCESKPEAEETCF DK ITYGETGGNSPVQEF TVPGSK NLQPASEYTVSLVAI K NTFAEVTGLSPGVTY YFK QDGHLWCSTTSNYE QDQK RPGGEPSPEGTTGQS YNQYSQR SSPVVIDASTAIDAPS NLR SYTITGLQPGTDYK TDELPQLVTLPHPNL HGPEILDVPSTVQK TNTNVNCPIECFMPL DVQADREDSRE VDVIPVNLPGEHGQR VTIMWTPPESAVTGY R VTWAPPPSIDLTNFL VR VVTPLSPPTNLHLEA NPDTGVLTVSWER WCGTTQNYDADQK AVGDKLPECEADDG CPKPPEIAHGYVEHS VR AVGDKLPECEAVCG KPK DIAPTLTLYVGK DYAEVGR GSFPWQAK LPECEADDGCPKPPEI AHGYVEHSVR LPECEAVCGKPK LRTEGDGVYTLNDK LRTEGDGVYTLNDK K LRTEGDGVYTLNNE K LRTEGDGVYTLNNE KQWINK QLVEIEK SCAVAEYGVYVK SPVGVQPILNEHTFC AGMSK TEGDGVYTLNDKKQ WINK TEGDGVYTLNNEK TEGDGVYTLNNEKQ WINK VGYVSGWGR VMPICLPSK VMPICLPSKDYAEVG R VTSIQDWVQK YVMLPVADQDQCIR 13 65 22 382 .45 50.6 -0.237 0.8 5 0.264 1.2 0 -0.357 0.7 8 P02774. 1 Vitamin D-binding protein 14 95 22 372 .13 51.2 -0.112 0.9 3 0.742 1.6 7 1.521 2.8 7 P01008. 1 Antithrom bin-III 47 EDFTSLSLVLYSR EFSHLGKEDFTSLSL VLYSR ELPEHTVK ELSSFIDK ELSSFIDKGQELCAD YSENTFTEYK ELSSFIDKGQELCAD YSENTFTEYKK EYANQFMWEYSTNY GQAPLSLLVSYTK GQELCADYSENTFTE YK HQPQEFPTYVEPTND EICEAFR HQPQEFPTYVEPTND EICEAFRK HQPQEFPTYVEPTND EICEAFRKDPK KELSSFIDKGQELCA DYSENTFTEYK LAQKVPTADLEDVLP LAEDITNILSK SCESNSPFPVHPGTAE CCTK SLGECCDVEDSTTCF NAK TAMDVFVCTYFMPA AQLPELPDVELPTNK THLPEVFLSK VCSQYAAYGEK VLEPTLK VMDKYTFELSR VPTADLEDVLPLAED ITNILSK YTFELSR ADGESCSASMMYQE GK AFLEVNEEGSEAAAS TAVVIAGR DDLYVSDAFHK DIPMNPMCIYR ELFYKADGESCSASM MYQEGK ELTPEVLQEWLDELE EMMLVVHMPR EQLQDMGLVDLFSPE K EVPLNTIIFMGR FRIEDGFSLKEQLQD MGLVDLFSPEK GDDITMVLILPKPEK IEDGFSLK ITDVIPSEAINELTVL VLVNTIYFK LPGIVAEGRDDLYVS DAFHK LQPLDFK LQPLDFKENAEQSR NDNDNIFLSPLSISTA FAMTK RVAEGTQVLELPFKG DDITMVLILPKPEK SKLPGIVAEGRDDLY VSDAFHK VAEGTQVLELPFK VAEGTQVLELPFKGD DITMVLILPKPEK VEKELTPEVLQEWLD ELEEMMLVVHMPR VWELSK 15 64 21 368 .56 43 -0.131 0.9 1 0.17 1.1 3 0.262 1.2 0 P00734. 2 Prothromb in 16 161 19 365 .85 46.7 0.203 1.1 5 1.627 3.0 9 1.794 3.4 7 P02790. 2 Hemopexi n 48 DKLAACLEGNCAEG LGTNYR ELLESYIDGR ENLDRDIALMK HQDFNSAVQLVENF CR ISMLEK ITDNMFCAGYKPDEG K ITDNMFCAGYKPDEG KR IVEGSDAEIGMSPWQ VMLFR KPVAFSDYIHPVCLP DRETAASLLQAGYK KSPQELLCGASLISDR LAACLEGNCAEGLG TNYR NPDSSTTGPWCYTTD PTVR RQECSIPVCGQDQVT VAMTPR SEGSSVNLSPPLEQC VPDR SEGSSVNLSPPLEQC VPDRGQQYQGR SGIECQLWR SPQELLCGASLISDR TATSEYQTFFNPR TFGSGEADCGLRPLF EK WYQMGIVSWGEGCD R WYQMGIVSWGEGCD RDGK DGWHSWPIAHQWPQ GPSAVDAAFSWEEK DVRDYFMPCPGR DYFMPCPGR EVGTPHGIILDSVDA AFICPGSSR EWFWDLATGTMK FDPVRGEVPPR GECQAEGVLFFQGD R GECQAEGVLFFQGD REWFWDLATGTMK GECQAEGVLFFQGD REWFWDLATGTMKE R GEFVWK LEKEVGTPHGIILDSV DAAFICPGSSR LLQDEFPGIPSPLDAA VECHR LWWLDLK NFPSPVDAAFR RLEKEVGTPHGIILDS VDAAFICPGSSR SGAQATWTELPWPH EK SGAQATWTELPWPH EKVDGALCMEK VDGALCMEK VWVYPPEK 17 71 19 336 .5 39.8 -0.285 0.8 2 0.728 1.6 6 0.892 1.8 6 P00747. 2 Plasminog en 18 72 17 320 .84 31.1 0.759 1.6 9 1.643 3.1 2 1.909 3.7 6 Q14624. 4 Interalphatrypsin inhibitor heavy chain H4 49 ATTVTGTPCQDWAA QEPHR DVVLFEK EAQLPVIENK FGMHFCGGTLISPEW VLTAAHCLEK FSPATHPSEGLEENY CR FVTWIEGVMR KLYDYCDVPQCAAP SFDCGKPQVEPK LFLEPTR LSSPAVITDK NPDADKGPWCFTTD PSVR NPDGDVGGPWCYTT NPR NPDNDPQGPWCYTT DPEKR TECFITGWGETQGTF GAGLLK TMSGLECQAWDSQS PHAHGYIPSK TPENYPNAGLTMNY CR VILGAHQEVNLEPHV QEIEVSR VIPACLPSPNYVVAD R VQSTELCAGHLAGG TDSCQGDSGGPLVCF EK WELCDIPR ATTVTGTPCQDWAA QEPHR DVVLFEK EAQLPVIENK FGMHFCGGTLISPEW VLTAAHCLEK FSPATHPSEGLEENY CR FVTWIEGVMR KLYDYCDVPQCAAP SFDCGKPQVEPK LFLEPTR LSSPAVITDK NPDADKGPWCFTTD PSVR NPDGDVGGPWCYTT NPR NPDNDPQGPWCYTT DPEKR TECFITGWGETQGTF GAGLLK TMSGLECQAWDSQS PHAHGYIPSK TPENYPNAGLTMNY CR VILGAHQEVNLEPHV QEIEVSR VIPACLPSPNYVVAD R VQSTELCAGHLAGG TDSCQGDSGGPLVCF EK WELCDIPR 19 53 17 274 .73 36.4 0.075 1.0 5 0.979 1.9 7 1.42 2.6 8 P01011. 2 Alpha-1antichymo trypsin 20 45 16 254 .97 45.4 -0.095 0.9 4 0.686 1.6 1 1.318 2.4 9 P06727. 3 Apolipopr otein A-IV 21 53 15 250 .09 36.3 -0.547 0.6 8 -0.227 0.8 5 -0.725 0.6 0 P04003. 2 C4bbinding protein alpha chain 50 ADLSGITGAR AKWEMPFDPQDTHQ SR AVLDVFEEGTEASAA TAVK AVLDVFEEGTEASAA TAVKITLLSALVETR DEELSCTVVELK EIGELYLPK EQLSLLDR EQLSLLDRFTEDAK GTHVDLGLASANVD FAFSLYK ITLLSALVETR LINDYVK LYGSEAFATDFQDSA AAK LYGSEAFATDFQDSA AAKK MEEVEAMLLPETLK MEEVEAMLLPETLK R RLYGSEAFATDFQDS AAAK WEMPFDPQDTHQSR ALVQQMEQLR ENADSLQASLRPHAD ELK IDQNVEELK IDQTVEELR LAPLAEDVR LEPYADQLR LGEVNTYAGDLQK LKEEIGKELEELR LTPYADEFK LTPYADEFKVK LVPFATELHER RVEPYGENFNKALV QQMEQLR SELTQQLNALFQDK SLAELGGHLDQQVEE FR SLAELGGHLDQQVEE FRR SLAPYAQDTQEKLN HQLEGLTFQMK CEWETPEGCEQVLTG K EDVYVVGTVLR FSAICQGDGTWSPR GVGWSHPLPQCEIVK KPDVSHGEMVSGFG PIYNYKDTIVFK LMQCLPNPEDVK LSLEIEQLELQR MALEVYK MALEVYKLSLEIEQL ELQR QSSSYSFFKEEIIYEC DK SHSTQTLTCNSDGEW VYNTFCIYK SRPANHCVYFYGDEI SFSCHETSR TPSCGDICNFPPK TWYPEVPKCEWETP EGCEQVLTGKR WTPYQGCEALCCPEP K 22 23 14 241 .85 14 1.381 2.6 0 2.044 4.1 2 2.917 7.5 5 P01031. 4 Complem ent C5 AFTECCVVASQLR ALVEGVDQLFTDYQI K DGHVILQLNSIPSSDF LCVR DVFLEMNIPYSVVR FSDASYQSINIPVTQN MVPSSR IDTQDIEASHYR IPLDLVPK LSMDIDVSYK MSAVEGICTSESPVID HQGTK MVETTAYALLTSLNL K QLPGGQNPVSYVYLE VVSK SYFPESWLWEVHLVP R TSTSEEVCSFYLK VSITSITVENVFVK 23 179 14 259 .83 56.6 0.125 1.0 9 0.165 1.1 2 -0.868 0.5 5 P01876. 2 Ig alpha-1 chain C region DASGVTFTWTPSSGK DLCGCYSVSSVLPGC AEPWNHGK GDTFSCMVGHEALPL AFTQK KGDTFSCMVGHEAL PLAFTQK LAGKPTHVNVSVVM AEVDGTCY NFPPSQDASGDLYTT SSQLTLPATQCLAGK QEPSQGTTTFAVTSIL R SGNTFRPEVHLLPPPS EELALNELVTLTCLA R TFTCTAAYPESK TPLTATLSK VAAEDWK VAAEDWKKGDTFSC MVGHEALPLAFTQK WLQGSQELPR WLQGSQELPREK 24 41 13 246 .37 25.9 -0.507 0.7 0 -1.449 0.3 7 -0.932 0.5 2 P00751. 2 Complem ent factor B ALFVSEEEK DAQYAPGYDKVKDI SEVVTPR EAGIPEFYDYDVALI K EELLPAQDIK EKLQDEDLGFL FLCTGGVSPYADPNT CR HVIILMTDGLHNMG GDPITVIDEIR KEAGIPEFYDYDVAL IK LLQEGQALEYVCPSG FYPYPVQTR QLNEINYEDHKLK VKDISEVVTPR VSEADSSNADWVTK WSGQTAICDNGAGY CSNPGIPIGTR 51 25 35 13 236 .8 44.9 0.472 1.3 9 1.611 3.0 5 1.783 3.4 4 P25311. 2 Zincalpha-2glycoprote in AGEVQEPELR AYLEEECPATLR DYIEFNK EDIFMETLK EIPAWVPFDPAAQIT K HVEDVPAFQALGSLN DLQFFR NILDRQDPPSVVVTS HQAPGEK QDPPSVVVTSHQAPG EK QKWEAEPVYVQR QVEGMEDWK QVEGMEDWKQDSQL QK WEAEPVYVQR YYYDGKDYIEFNK 26 55 12 212 .34 23.8 -0.245 0.8 4 -0.446 0.7 3 0.278 1.2 1 P19823. 2 Interalphatrypsin inhibitor heavy chain H2 27 34 12 205 .15 20.9 0.38 1.3 0 0.389 1.3 1 0.624 1.5 4 P19827. 3 Interalphatrypsin inhibitor heavy chain H1 28 73 11 199 .84 36.5 -1.059 0.4 8 0.678 1.6 0 0.322 1.2 5 P01871. 3 Ig mu chain C region AEDHFSVIDFNQNIR ETAVDGELVVLYDV KREEK FLHVPDTFEGHFDGV PVISK HLEVDVWVIEPQGLR IQPSGGTNINEALLR MLADAPPQDPSCCSG ALYYGSK NVQFNYPHTSVTDVT QNNFHNYFGGSEIVV AGK SILQMSLDHHIVTPLT SLVIENEAGDER SLPGESEEMMEEVDQ VTLYSYKVQSTITSR SSALDMENFR TILDDLR TILDDLRAEDHFSVID FNQNIR ADVQAHGEGQEFSIT CLVDEEEMK ADVQAHGEGQEFSIT CLVDEEEMKK EVAFDLEIPK GMADQDGLKPTIDK PSEDSPPLEMLGPR GSLVQASEANLQAA QDFVR LDAQASFLPK LWAYLTIQELLAK QAVDTAVDGVFIR QLVHHFEIDVDIFEPQ GISK QYYEGSEIVVAGR TAFISDFAVTADGNA FIGDIKDKVTAWK VTFQLTYEEVLK DVMQGTDEHVVCK EGKQVGSGVTTDQV QAEAK ESATITCLVTGFSPAD VFVQWMQR FTCTVTHTDLPSPLK GQPLSPEKYVTSAPM PEPQAPGR GVALHRPDVYLLPPA R NVPLPVIAELPPK QVGSGVTTDQVQAE AK STGKPTLYNVSLVMS DTAGTCY YAATSQVLLPSKDV MQGTDEHVVCK YVTSAPMPEPQAPGR 52 29 49 11 184 .25 21.1 -0.317 0.8 0 0.306 1.2 4 0.19 1.1 4 P01042. 2 Kininogen -1 DFVQPPTK DIPTNSPELEETLTHTI TK ENFLFLTPDCK FKLDDDLEHQGGHV LDHGHK IASFSQNCDIYPGKDF VQPPTK LGQSLDCNAEVYVV PWEK TVGSDTFYSFK TVGSDTFYSFKYEIK TWQDCEYKDAAK YEIKEGDCPVQSGK YFIDFVAR 30 16 11 156 .93 16.7 0.145 1.1 1 1.876 3.6 7 1.754 3.3 7 P04264. 6 Keratin, type II cytoskelet al 1 DYQELMNTK IEISELNR NMQDMVEDYR QISNLQQSISDAEQR SKAEAESLYQSK SKAEAESLYQSKYEE LQITAGR SLDLDSIIAEVK SLDLDSIIAEVKAQYE DIAQK TNAENEFVTIK WELLQQVDTSTR YEELQITAGR 31 72 10 204 .75 78.2 1.339 2.5 3 4.294 19. 62 3.083 8.4 7 P68871. 2 Hemoglob in subunit beta 32 25 10 189 .08 26.7 -0.989 0.5 0 -1.061 0.4 8 -0.561 0.6 8 P06396. 1 Gelsolin EFTPPVQAAYQK FFESFGDLSTPDAVM GNPK FFESFGDLSTPDAVm GNPK GTFATLSELHCDK GTFATLSELHCDKLH VDPENFR LHVDPENFR LLVVYPWTQR SAVTALWGK VLGAFSDGLAHLDN LK VNVDEVGGEALGR VVAGVANALAHKYH AQPVQVAEGSEPDGF WEALGGK DSQEEEKTEALTSAK EVQGFESATFLGYFK NWRDPDQTDGLGLS YLSSHIANVER QTQVSVLPEGGETPL FK SEDCFILDHGK TPSAAYLWVGTGAS EAEK VHVSEEGTEPEAMLQ VLGPKPALPAGTEDT AK VPEARPNSMVVEHPE FLK VPFDAATLHTSTAM AAQHGMDDDGTGQ K 53 33 23 9 169 .19 21 0.722 1.6 5 0.873 1.8 3 1.682 3.2 1 P43652. 1 Afamin 34 19 9 154 .97 31.7 0.528 1.4 4 1.256 2.3 9 1.746 3.3 5 P08697. 3 Alpha-2antiplasmi n 35 37 8 164 .39 34.8 -0.721 0.6 1 -0.451 0.7 3 -0.717 0.6 1 P02765. 1 Alpha-2HSglycoprote in 36 20 8 153 .51 20.4 0.08 1.0 6 0.422 1.3 4 0.632 1.5 5 P10643. 2 Complem ent componen t C7 37 30 8 156 .9 36.8 -0.053 0.9 6 2.741 6.6 9 2.19 4.5 6 P02763. 1 Alpha-1acid glycoprote in 1 /orosomuc oid 1 54 AESPEVCFNEESPK ESLLNHFLYEVAR HELTDEELQSLFTNF ANVVDK HPDLSIPELLR IAPQLSTEELVSLGEK KSDVGFLPPFPTLDPE EK LKHELTDEELQSLFT NFANVVDK SDVGFLPPFPTLDPEE K TYVPPPFSQDLFTFH ADMCQSQNEELQR DSFHLDEQFTVPVEM MQAR ELKEQQDSPGNKDFL QSLK EQQDSPGNKDFLQSL K GFPIKEDFLEQSEQLF GAKPVSLTGK HQMDLVATLSQLGL QELFQAPDLR IQEFLSGLPEDTVLLL LNAIHFQGFWR LCQDLGPGAFR QEDDLANINQWVK WFLLEQPEIQVAHFP FK AQLVPLPPSTYVEFT VSGTDCVAK EHAVEGDCDFQLLK EHAVEGDCDFQLLK LDGK HTFMGVVSLGSPSGE VSHPR TVVQPSVGAAAGPV VPPCPGR VWPQQPSGELFEIEID TLETTCHVLDPTPVA R ELSHLPSLYDYSAYR GGGAGFISGLSYLEL DNPAGNKR LLEPHCFPLSLVPTEF CPSPPALK LTPLYELVK SCVGETTESTQCEDE ELEHLR SVAVYGQYGGQPCV GNAFETQSCEPTR VLFYVDSEK VTVSCSGGMSLEGPS AFLCGSSLK EQLGEFYEALDCLR NWGLSVYADKPETT K SDVVYTDWK SDVVYTDWKK TEDTIFLR TYMLAFDVNDEK TYMLAFDVNDEKNW GLSVYADKPETTK YVGGQEHFAHLLILR 38 23 8 143 .68 35.6 1.318 2.4 9 0.562 1.4 8 1.019 2.0 3 P02649. 1 Apolipopr otein E 39 22 8 143 .1 30.8 -0.857 0.5 5 -0.4 0.7 6 -0.68 0.6 2 O43866. 1 CD5 antigenlike 40 14 8 134 .58 22.4 0.099 1.0 7 0.982 1.9 8 1.68 3.2 0 P36955. 4 Pigment epithelium -derived factor 41 42 8 133 .48 31.8 0.066 1.0 5 0.375 1.3 0 0.436 1.3 5 P02749. 3 Beta-2glycoprote in 1 42 25 8 129 .38 23.6 0.119 1.0 9 0.238 1.1 8 0.839 1.7 9 P10909. 1 Clusterin 43 48 7 152 .35 72.7 0.207 1.1 5 0.509 1.4 2 0.892 1.8 6 P02766. 1 Transthyre tin 55 AATVGSLAGQPLQER ALMDETMKELK AYKSELEEQLTPVAE ETR FWDYLR GEVQAMLGQSTEEL R SWFEPLVEDMQR VQAAVGTSAAPVPS DNH WVQTLSEQVQEELLS SQVTQELR EATLQDCPSGPWGK ELGCGAASGTPSGIL YEPPAEK ELGCGAASGTPSGIL YEPPAEKEQK FWGFHDCTHQEDVA VICSG GQWGTVCDDGWDIK GQWGTVCDDGWDIK DVAVLCR GVWGSVCDDNWGE KEDQVVCK IWLDNVR ALYYDLISSPDIHGTY K ALYYDLISSPDIHGTY KELLDTVTAPQK EIPDEISILLLGVAHF K ELLDTVTAPQK KTSLEDFYLDEER LAAAVSNFGYDLYR LQSLFDSPDFSK YGLDSDLSCK ATFGCHDGYSLDGPE EIECTK CSYTEDAQCIDGTIE VPK CTEEGKWSPELPVCA PIICPPPSIPTFATLR DKATFGCHDGYSLD GPEEIECTK KCSYTEDAQCIDGTI EVPK TCPKPDDLPFSTVVP LK TFYEPGEEITYSCKPG YVSR WSPELPVCAPIICPPP SIPTFATLR ASSIIDELFQDR EILSVDCSTNNPSQA K KTLLSNLEEAK LFDSDPITVTVPVEVS R QQTHMLDVMQDHFS R RELDESLQVAER TLLSNLEEAK VTTVASHTSDSDVPS GVTEVVVK AADDTWEPFASGK ALGISPFHEHAEVVF TANDSGPR GSPAINVAVHVFR KAADDTWEPFASGK TSESGELHGLTTEEEF VEGIYK TSESGELHGLTTEEEF VEGIYKVEIDTK YTIAALLSPYSYSTTA VVTNPKE 44 34 7 128 .73 23.8 -0.213 0.8 6 0.175 1.1 3 0.022 1.0 2 P04217. 4 Alpha-1Bglycoprote in 45 20 7 120 .08 13.8 0.698 1.6 2 2.23 4.6 9 2.817 7.0 5 P05155. 2 Plasma protease C1 inhibitor 46 50 7 112 .67 85.8 0.356 1.2 8 1.266 2.4 0 0.362 1.2 9 P0CG05 .1 Ig lambda2 chain C regions 47 12 7 111 .61 11.5 -0.227 0.8 5 1.483 2.8 0 1.061 2.0 9 P13671. 3 Complem ent componen t C6 48 27 7 108 .78 19.2 -0.396 0.7 6 -0.19 0.8 8 0.219 1.1 6 P04004. 1 Vitronecti n 49 26 7 105 .71 49.2 0.778 1.7 1 2.857 7.2 5 1.029 2.0 4 P69905. 2 Hemoglob in subunit alpha 56 CEGPIPDVTFELLR LELHVDGPPPRPQLR LLELTGPK SLPAPWLSMAPVSWI TPGLK SWVPHTFESELSDPV ELLVAES TPGAAANLELIFVGP QHAGNYR VTLTCVAPLSGVDFQ LR GVTSVSQIFHSPDLAI R IKVTTSQDMLSIMEK LEDMEQALSPSVFK LLDSLPSDTR TNLESILSYPK VTTSQDMLSIMEK AAPSVTLFPPSSEELQ ANK ADSSPVKAGVETTTP SK AGVETTTPSK AGVETTTPSKQSNNK YAASSYLSLTPEQWK ATLVCLISDFYPGAV TVAWK SYSCQVTHEGSTVEK YAASSYLSLTPEQWK DLTSLGHNENQQGSF SSQGGSSFSVPIFYSS K IGESIELTCPK SEYGAALAWEK TECIKPVVQEVLTITP FQR TFSEWLESVK VPANLENVGFEVQT AEDDLK VPANLENVGFEVQT AEDDLKTDFYK DWHGVPGQVDAAM AGR GQYCYELDEK LIRDVWGIEGPIDAAF TR MDWLVPATCEPIQSV FFFSGDK RVDTVDPPYPR SIAQYWLGCPAPGHL VDTVDPPYPR KVADALTNAVAHVD DMPNALSALSDLHA HK KVADALTNAVAHVD DMPNALSALSDLHA HKLR TYFPHFDLSHGSAQV K VADALTNAVAHVDD MPNALSALSDLHAH K VADALTNAVAHVDD MPNALSALSDLHAH KLR VDPVNFK VGAHAGEYGAEALE R 50 22 6 114 .22 30.3 0.563 1.4 8 3.444 10. 88 3.097 8.5 6 P19652. 2 Alpha-1acid glycoprote in 2 51 23 6 110 .95 21.5 -0.121 0.9 2 0.276 1.2 1 0.334 1.2 6 P02760. 1 Protein AMBP 52 12 6 92. 51 26.4 -0.779 0.5 8 -0.471 0.7 2 0.414 1.3 3 P27169. 3 Serum paraoxona se/aryleste rase 1 53 52 5 93. 68 79.2 0.006 1.0 0 1.529 2.8 9 0.753 1.6 9 P01834. 1 Ig kappa chain C region 54 9 5 85. 93 14 -0.264 0.8 3 0.869 1.8 3 -0.298 0.8 1 P07357. 2 Complem ent componen t C8 alpha chain 55 12 5 72. 06 10.2 -0.211 0.8 6 0.308 1.2 4 0.186 1.1 4 P05156. 2 Complem ent factor I AQLGDLPWQVAIK GLETSLAECTFTK IVIEYVDR TMGYQDFADVVCYT QK VFSLQWGEVK 56 9 5 71. 22 10.5 -0.022 0.9 8 -0.283 0.8 2 0.613 1.5 3 P06681. 2 Complem ent C2 57 7 5 69. 93 13.7 1.051 2.0 7 1.987 3.9 6 1.562 2.9 5 P08670. 2 Vimentin precursor AVISPGFDVFAK KNQGILEFYGDDIAL LK LDVDWR LLGMETMAWQEIR QPYSYDFPEDVAPAL GTSFSHMLGATNPTQ K MSTRSVSSSS EEMLQREEAE RKLLEGEESR ISLPLPNFSS TLLIKTVETR 57 EQLGEFYEALDCLCI PR NWGLSFYADKPETT K SDVMYTDWK TEDTIFLR TLMFGSYLDDEK TLMFGSYLDDEKNW GLSFYADKPETTK AFIQLWAFDAVK EDSCQLGYSAGPCM GMTSR ETLLQDFR GECVPGEQEPEPILIP R KEDSCQLGYSAGPC MGMTSR VVAQGVGIPEDSIFT MADR EVQPVELPNCNLVK FDVSSFNPHGISTFTD EDNAMYLLVVNHPD AK GIETGSEDLEILPNGL AFISSGLKYPGIK IFFYDSENPPASEVLR ILLMDLNEEDPTVLE LGITGSK IQNILTEEPK DSTYSLSSTLTLSK SGTASVVCLLNNFYP R TVAAPSVFIFPPSDEQ LK VDNALQSGNSQESVT EQDSK VYACEVTHQGLSSPV TK ALDQYLMEFNACR AMAVEDIISR FGGTICSGDIWDQAS CSSSTTCVR LGSLGAACEQTQTEG AK YNPVVIDFEMQPIHE VLR 58 7 5 69. 14 14.6 1.098 2.1 4 2.895 7.4 4 3.015 8.0 8 P05546. 3 Heparin cofactor 2 GGETAQSADPQWEQ LNNK HQGTITVNEEGTQAT TVTTVGFMPLSTQVR IAIDLFK NYNLVESLK QFPILLDFK 59 36 5 73. 15 37.3 0.216 1.1 6 0.759 1.6 9 1.834 3.5 7 P02656. 1 Apolipopr otein C-III 60 32 4 89. 49 9.8 0.117 1.0 8 1.82 3.5 3 2.138 4.4 0 P01019. 1 Angiotens inogen DALSSVQESQVAQQ AR DYWSTVK GWVTDGFSSLK GWVTDGFSSLKDYW STVK TAKDALSSVQESQVA QQAR ALQDQLVLVAAK DPTFIPAPIQAK SLDFTELDVAAEK SLDFTELDVAAEKID R 61 22 4 76. 36 12.3 1.212 2.3 2 1.145 2.2 1 0.544 1.4 6 P04196. 1 Histidinerich glycoprote in DGYLFQLLR GEVLPLPEANFPSFPL PHHK GGEGTGYFVDFSVR YKEENDDFASFRVDR 62 5 4 71. 51 11.7 -0.061 0.9 6 1.442 2.7 2 1.427 2.6 9 P35908. 2 Keratin, type II cytoskelet al 2 epidermal IEISELNR NLDLDSIIAEVK SKEEAEALYHSKYEE LQVTVGR VDLLNQEIEFLK 63 42 4 71. 67 42 0.95 1.9 3 3.061 8.3 5 3.757 13. 52 P02652. 1 Apolipopr otein A-II 64 9 4 71. 13 14.4 -0.588 0.6 7 -1.253 0.4 2 -0.359 0.7 8 Q96PD5 .1 Nacetylmur amoyl-Lalanine amidase 65 11 4 69. 71 17.7 0.279 1.2 1 1.562 2.9 5 2.39 5.2 4 P08185. 1 Corticoste roidbinding globulin EPCVESLVSQYFQTV TDYGK EPCVESLVSQYFQTV TDYGKDLMEK EPCVESLVSQYFQTV TDYGKDLmEK EQLTPLIK SPELQAEAK EFTEAFLGCPAIHPR EGKEYGVVLAPDGS TVAVEPLLAGLEAGL QGR GCPDVQASLPDAK GSQTQSHPDLGTEGC WDQLSAPR GTWTQPFDLASTR HYYESEVLAMNFQD WATASR IVDLFSGLDSPAILVL VNYIFFK WSAGLTSSQVDLYIP K 66 8 4 68. 08 9.7 0.486 1.4 0 2.058 4.1 6 2.259 4.7 9 P13645. 6 Keratin, type I cytoskelet al 10 IEISELNR NLDLDSIIAEVK SKEEAEALYHSKYEE LQVTVGR VDLLNQEIEFLK 67 8 4 66. 79 23.2 -0.295 0.8 2 0.641 1.5 6 1.573 2.9 8 P05090. 1 Apolipopr otein D CPNPPVQENFDVNK MTVTDQVNCPK NPNLPPETVDSLK WYEIEK 58 68 8 4 65. 72 43.5 0.582 1.5 0 0.618 1.5 3 1.72 3.2 9 P02655. 1 Apolipopr otein C-II ESLSSYWESAK STAAMSTYTGIFTDQ VLSVLK STAAMSTYTGIFTDQ VLSVLKGEE TYLPAVDEK 69 10 3 68. 38 14.4 -0.056 0.9 6 0.839 1.7 9 1.445 2.7 2 P35527. 3 Keratin, type I cytoskelet al 9 MTLDDFR SDLEMQYETLQEEL MALK TLLDIDNTR 70 7 3 61. 94 11.6 0.233 1.1 8 -0.402 0.7 6 0.018 1.0 1 P02748. 2 Complem ent componen t C9 AIEDYINEFSVR FEGIACEISK RQCVPTEPCEDAEDD CGNDFQCSTGR 71 7 3 54. 01 16.8 -0.514 0.7 0 -0.74 0.6 0 -0.604 0.6 6 P07360. 3 72 6 3 51. 01 8.2 -0.094 0.9 4 0.36 1.2 8 0.782 1.7 2 P07358. 3 VQEAHLTEDQIFYFP K YGFCEAADQFHVLD EVR YGFCEAADQFHVLD EVRR DTMVEDLVVLVR IPGIFELGISSQSDR VKVEPLYELVTATDF AYSSTVR 73 5 3 50. 09 14.8 -1.661 0.3 2 -0.514 0.7 0 -1.986 0.2 5 P01880. 2 Complem ent componen t C8 gamma chain Complem ent componen t C8 beta chain Ig delta chain C region 74 4 3 49. 63 8.1 -0.145 0.9 0 -0.237 0.8 5 0.724 1.6 5 P09871. 1 Complem ent C1s subcompo nent LLEVPEGR MGPTVSPICLPGTSSD YNLMDGDLGLISGW GR SNALDIIFQTDLTGQK 75 5 3 48. 97 13.6 -0.486 0.7 1 -0.024 0.9 8 0.426 1.3 4 P51884. 2 Lumican NIPTVNENLENYYLE VNQLEK SLEDLQLTHNK SLEYLDLSFNQIAR 76 3 3 46. 74 5.6 -0.455 0.7 3 0.587 1.5 0 0.876 1.8 4 P07225. 1 Vitamin Kdependent protein S FSAEFDFR NIPGDFECECPEGYR SQDILLSVENTVIYR 77 5 3 44. 46 6.1 0.288 1.2 2 -0.01 0.9 9 0.103 1.0 7 P03952. 1 Plasma kallikrein GGDVASMYTPNAQY CQMR VAEYMDWILEK VLTPDAFVCR 78 3 3 44. 16 9.1 0.459 1.3 7 1.226 2.3 4 1.276 2.4 2 P00748. 3 Coagulati on factor XII AEEHTVVLTVTGEPC HFPFQYHR LHEAFSPVSYQHDLA LLR NPDNDIRPWCFVLNR 79 3 3 39. 68 8.3 -0.46 0.7 3 -0.065 0.9 6 0.874 1.8 3 P05154. 3 Plasma serine protease inhibitor ADFTFDLYR GFQQLLQELNQPR MQILEGLGLNLQK 80 5 3 34. 62 17.8 0.89 1.8 5 1.503 2.8 3 0.264 1.2 0 P20851. 1 C4bbinding protein beta chain EVEGQILGTYVCIK NLCEAMENFMQQLK SQCLEDHTWAPPFPI CK 59 DSYYMTSSQLSTPLQ QWR TPECPSHTQPLGVYL LTPAVQDLWLR VPTGGVEEGLLER 81 8 2 69. 93 12.8 0.121 1.0 9 0.73 1.6 6 0.649 1.5 7 P05160. 3 Coagulati on factor XIII B chain Complem ent factor H-related protein 2 Ig kappa chain VIII region GOL Ficolin-3 GDTYPAELYITGSILR WSSPPVCLEPCTVNV DYMNR 82 8 2 49. 53 17.4 0.155 1.1 1 0.649 1.5 7 0.482 1.4 0 P36980. 1 83 14 2 41. 95 30.2 -0.459 0.7 3 -0.079 0.9 5 -0.326 0.8 0 P04206. 1 84 6 2 40. 28 13.7 -0.296 0.8 1 -0.831 0.5 6 0.79 1.7 3 O75636. 2 85 7 2 40. 02 7.1 0.303 1.2 3 -2.501 0.1 8 -1.798 0.2 9 P02743. 2 Serum amyloid Pcomponen t Ig kappa chain V-II region TEW IVLGQEQDSYGGK IVLGQEQDSYGGKFD R 86 8 2 38. 38 31.8 -1.137 0.4 5 0.201 1.1 5 -0.192 0.8 8 P01617. 1 87 4 2 35. 37 10.9 -1.378 0.3 8 -0.897 0.5 4 -0.822 0.5 7 P01857. 1 Ig gamma1 chain C region 2.4 7 P05543. 2 Thyroxine -binding globulin TPEVTCVVVDVSHED PEVK TTPPVLDSDGSFFLYS K EGQMESVEAAMSSK GTEAAAVPEVELSDQ PENTFLHPIIQIDR 88 5 2 34. 94 10.3 0.108 1.0 8 1.05 2.0 7 1.304 89 3 2 32. 73 7 0.362 1.2 9 1.105 2.1 5 1.99 3.9 7 P29622. 3 Kallistatin EIEEVLTPEMLMR VPMMLQDQEHHWY LHDR 90 4 2 30. 17 6.5 0.27 1.2 1 -0.58 0.6 7 0.363 1.2 9 O14791. 5 Apolipopr otein L1 VNEPSILEMSR VTEPISAESGEQVER 91 2 2 29. 91 5.7 -0.626 0.6 5 -0.994 0.5 0 -0.959 0.5 1 Q9BXR 6.1 Complem ent factor H-related protein 5 EIMENYNIALR ITCTEEGWSPTPK YKPFSQVPTGEVFYY SCEYNFVSPSK 92 3 2 28. 95 7.1 0.658 1.5 8 0.94 1.9 2 1.342 2.5 4 Q14520. 1 Plasma hyalurona n-binding protein NPDADEKPWCFIK TVCLPDGSFPSGSEC HISGWGVTETGK 93 4 2 27. 77 18.7 0.134 1.1 0 -0.338 0.7 9 0.397 1.3 2 P02775. 3 GKEESLDSDLYAELR ICLDPDAPR -0.002 1.0 0 0.063 1.0 4 0.605 1.5 2 P35858. 1 Platelet basic protein Insulinlike growth factorbinding protein complex acid labile subunit 94 2 2 27. 67 4.6 60 TGDIVEFVCK YKPFSQVPTGEVFYY SCEYNFVSPSK EIVLTQSPGTLSLSPG ER FSGSGSGTDFTLTISR ALPVFCDMDTEGGG WLVFQR ELLSQGATLSGWYH LCLPEGR DIVMTQSPLSLPVTP GEPASISCR FSGSGSGTDFTLK DLHFLEELQLGHNR LAELPADALGPLQR 95 4 2 27. 28 4.7 -0.618 0.6 5 0.087 1.0 6 0.173 1.1 3 P22792. 3 Carboxyp eptidase N subunit 2 AGGSWDLAVQER DHLGFQVTWPDESK 96 5 2 25. 48 8.8 0.369 1.2 9 1.276 2.4 2 1.551 2.9 3 P22352. 2 FLVGPDGIPIMR MDILSYMR 97 5 1 32. 08 7.4 1.574 2.9 8 3.349 10. 19 2.292 4.9 0 P02750. 2 98 3 1 28. 77 4.9 -0.662 0.6 3 -0.354 0.7 8 -0.007 1.0 0 Q06033. 2 99 3 1 25 5.7 0.297 1.2 3 -0.413 0.7 5 0.302 1.2 3 Q96KN 2.4 10 0 12 1 23. 86 15 0.485 1.4 0 0.12 1.0 9 -0.172 0.8 9 P01766. 1 10 1 4 1 22. 6 15.7 -0.303 0.8 1 -0.294 0.8 2 -0.46 0.7 3 P01598. 1 10 2 6 1 20. 6 15.7 -0.303 0.8 1 -0.017 0.9 9 -0.511 0.7 0 P01593. 1 10 3 1 1 20. 58 2.9 -0.997 0.5 0 -0.01 0.9 9 1.064 2.0 9 P80108. 3 10 4 1 1 20. 35 2.3 0.24 1.1 8 0.751 1.6 8 0.047 1.0 3 P02671. 2 10 5 1 1 20. 17 15.7 -0.466 0.7 2 0.175 1.1 3 -0.303 0.8 1 P01611. 1 10 6 10 7 10 8 2 1 0.2 -0.779 3.4 -1.914 6 1 15.1 -0.452 1.0 6 0.8 2 1.5 2 -1.914 1 0.5 8 0.2 7 0.7 3 0.078 1 19. 01 18. 97 18. 57 0.2 7 0.5 5 0.9 2 P08519. 1 P06276. 1 P01774. 1 10 9 2 1 18. 3 14.9 -2.438 0.1 8 -0.424 0.7 5 -0.6 0.6 6 P01625. 2 11 0 5 1 17. 59 9.4 -0.465 0.7 2 -0.294 0.8 2 -0.165 0.8 9 P01591. 4 11 1 2 1 17. 31 15.7 0.32 1.2 5 -1.093 0.4 7 0.113 1.0 8 P01714. 1 11 2 1 1 16. 92 3.9 0 1.0 0 0 1.0 0 0 1.0 0 P01854. 1 11 3 11 4 2 1 10.8 0.593 16.8 1.542 39. 84 0.5 8 5.85 1 1.5 1 2.9 1 5.316 3 16. 69 16. 17 57. 68 0.8 2 P02654. 1 P01717. 1 Glutathion e peroxidase 3 Leucinerich alpha2glycoprote in Interalphatrypsin inhibitor heavy chain H3 Beta-AlaHis dipeptidas e Ig heavy chain VIII region BRO Ig kappa chain V-I region EU Ig kappa chain V-I region AG Phosphati dylinositol -glycanspecific phospholi pase D Fibrinoge n alpha chain Ig kappa chain V-I region Wes Apolipopr otein(a) Cholineste rase Ig heavy chain VIII region POM Ig kappa chain VIV region Len Immunogl obulin J chain Ig lambda chain VIII region SH Ig epsilon chain C region Apolipopr otein C-I Ig lambda chain VIV region -0.28 0.608 -0.776 61 -0.859 -0.115 -0.286 DLLLPQPDLR STSIVIMLTDGDANV GESRPEK AIHLDLEEYR EVQLVESGGGLVQP GGSLR DIQMTQSPSTLSASV GDR DIQMTQSPSSLSASV GDR FGGVLHLSDLDDDG LDEIIMAAPLR TFPGFFSPMLGEFVSE TESR DIQMTQSPSSVSASV GDR EWFSETFQK AILQSGSFNAPWAVT SLYEAR EVQLLESGGGLVQPG GSL DIVMTQSPDSLAVSL GER CYTAVVPLVYGGET K SELTQDPAVSVALGQ TVR AAPEVYAFATPEWP GS WIYHLTEGSTDLR SYELTQPPSVSVSPG QTAR Hil 11 5 2 1 16. 03 4.8 -0.731 0.6 0 -0.426 0.7 4 0.059 1.0 4 P15169. 1 11 6 2 1 15. 76 16.3 1.213 2.3 2 1.303 2.4 7 1.018 2.0 3 P0DJI8. 1 11 7 2 1 15. 68 3.7 -0.355 0.7 8 0.782 1.7 2 0.888 1.8 5 Q96IY4 .2 11 8 1 1 15. 59 7.8 -0.601 0.6 6 -0.353 0.7 8 -1.48 0.3 6 P04433. 1 11 9 1 1 15. 46 20.5 -3.216 0.1 1 -3.216 0.1 1 0 1.0 0 P01616. 1 12 0 12 1 12 2 1 1 1 -0.79 3.6 -0.633 1 1 5.1 -0.768 0.4 6 1.1 9 1.1 3 -1.058 1 0.5 8 0.6 4 0.5 9 -1.11 2 15. 22 15. 18 14. 72 0.4 8 1.6 0 2.6 2 P07996. 2 P27918. 2 P01861. 1 12 3 1 1 14 5.2 -1.191 0.4 4 -0.393 0.7 6 -2.173 0.2 2 P01859. 2 12 4 12 5 12 6 1 1 11.8 1.079 10.8 -0.316 1 1 11.5 0.476 1.7 5 3.1 3 0.2 5 1.519 1 2.1 1 0.8 0 1.3 9 0.809 1 13. 71 13. 49 13. 47 2.8 7 4.4 1 0.3 1 P05109. 1 P31151. 4 P35542. 2 12 7 4 1 13. 33 4.5 -1.809 0.2 9 -4.079 0.0 6 -4.277 0.0 5 P50897. 1 12 8 2 1 13. 27 7.7 1.182 2.2 7 3.267 9.6 3 2.284 4.8 7 P01781. 1 12 9 3 1 13. 05 2.7 -0.041 0.9 7 2.091 4.2 6 3.573 11. 90 P50213. 1 13 0 13 1 2 1 6.9 -0.502 15.7 -0.377 2.2 0 0.3 1 1.853 1 0.7 1 0.7 7 1.137 1 12. 97 12. 74 3.6 1 0.2 2 O95445. 2 P04430. 1 13 2 1 1 12. 6 7.5 -0.26 0.8 4 -1.288 0.4 1 0.82 1.7 7 P04070. 1 13 3 13 4 1 1 13.1 1.091 0.6 -1.459 22. 75 0.3 5 4.485 1 2.1 3 0.3 6 4.508 1 12. 39 12. 32 22. 39 0.4 6 P06702. 1 Q5XPI4 .1 13 5 1 1 11. 84 4.5 -0.161 1.0 1 P00742. 2 0.8 9 0.248 0.173 1.645 -1.972 -1.688 -1.528 -1.043 0.4 9 62 0.675 1.389 2.141 -1.698 -2.158 -1.132 0.017 Carboxyp eptidase N catalytic chain Serum amyloid A-1 protein Carboxyp eptidase B2 Ig kappa chain VIII region VG Ig kappa chain V-II region MIL Thrombos pondin-1 Properdin Ig gamma4 chain C region Ig gamma2 chain C region Protein S100-A8 Protein S100-A7 Serum amyloid A-4 protein Palmitoylprotein thioesteras e1 Ig heavy chain VIII region GAL Isocitrate dehydroge nase [NAD] subunit alpha, mitochond rial Apolipopr otein M Ig kappa chain V-I region BAN Vitamin Kdependent protein C Protein S100-A9 E3 ubiquitinprotein ligase RNF123 Coagulati on factor HLYVLEFSDHPGIHE PLEPEV FFGHGAEDSLADQA ANEWG * DIQMTQSPSSLSASV GDR DIVLTQSPLSLPVTPG EPASISCR FVFGTTPEDILR * TTPPVLDSDGSFFLYS R TTPPMLDSDGSFFLY SK LLETECPQYIR GTNYLADVFEK SFFKEALQGVGDMG R CPGESSHICDFIRK GLEWVANIK IAEFAFEYAR WIYHLTEGSTDLR DIQLTQSPSSLSASVG DR STTDNDIALLHLAQP ATLSQTIVPICLPDSG LAER LLETECPQYIR FLQENASGR ITVVAGEHNIEETEHT EQKR X 13 6 13 7 1 1 11. 6 11. 49 2.3 -0.465 1 1 13 8 13 9 2 1 1 1 14 0 1 14 1 0.7 2 0.5 8 7.2 6 0.2 8 P43251. 2 P00740. 2 4.3 -0.789 11. 36 11. 26 1.5 0.306 1.3 1 0.9 8 O15265. 1 Q7Z353 .1 3.9 -0.606 1 11. 23 2.4 1.952 3.8 7 -1.295 0.4 1 2.614 6.1 2 P01833. 4 2 1 11. 16 10.9 -1.142 0.4 5 -3.046 0.1 2 -4.978 0.0 3 Q6ZN0 3.1 14 2 1 1 11. 14 0.4 0.117 1.0 8 0 1.0 0 -1.596 0.3 3 Q96M8 6.2 14 3 1 1 11. 13 4.7 -0.191 0.8 8 -2.062 0.2 4 -1.817 0.2 8 Q8N336 .3 14 4 14 5 2 1 5.2 0.039 1 1 11. 12 10. 91 1.0 3 0.6 2 1.466 2.7 6 0.5 7 1.978 3.9 4 0.6 8 P09486. 1 Q04756. 1 2.9 -0.682 14 6 1 1 10. 89 11 2.137 4.4 0 3.163 8.9 6 0.979 1.9 7 P02745. 2 14 7 1 1 10. 69 2 0.828 1.7 8 3.24 9.4 5 3.489 11. 23 Q9C0A 0.3 14 8 1 1 10. 54 4.5 2.535 5.8 0 5.277 38. 77 3.418 10. 69 P00915. 2 14 9 1 1 10. 51 0.7 -0.839 0.5 6 -0.232 0.8 5 -0.346 0.7 9 Q9P2R6 .2 15 0 15 1 1 1 1 1 10. 47 10. 3 6.9 0 0 1.345 1.0 0 1.3 2 0 11.8 1.0 0 2.5 4 1.0 0 1.0 0 P05452. 3 Q16514. 1 15 2 1 1 10. 27 6.4 0.952 1.9 3 1.558 2.9 4 1.605 3.0 4 Q9HBK 9.3 15 3 2 1 10. 21 1.3 0.085 1.0 6 -3.562 0.0 8 -4.584 0.0 4 Q8WV M7.3 15 4 1 1 10. 21 0.6 -0.74 0.6 0 -0.598 0.6 6 -0.877 0.5 4 Q9P1Q0 .2 1.2 4 0.6 6 2.534 -2.115 0.67 0.643 -0.815 0.399 5.7 9 0.2 3 1.5 9 1.5 6 63 2.859 -1.821 0.39 -0.032 -0.56 0 Biotinidas e Coagulati on factor IX Ataxin-7 Highly divergent homeobox Polymeric immunogl obulin receptor Uncharact erized protein encoded by LINC0032 2 Dynein heavy chain domaincontaining protein 1 ELMO domaincontaining protein 1 SPARC Hepatocyt e growth factor activator Complem ent C1q subcompo nent subunit A Contactinassociated proteinlike 4 Carbonic anhydrase 1 Arginineglutamic acid dipeptide repeats protein Tetranecti n Transcript ion initiation factor TFIID subunit 12 Arsenite methyltran sferase Cohesin subunit SA-1 Vacuolar protein sorting- VDLITFDTPFAGR ITVVAGEHNIEETEHT EQKR IPPVPSTTSPISTR KNYGNSSVQASEMT VPQKPSVCHRPCK NADLQVLKPEPELVY EDLR * * GMGLLGLYNLQYFA ER YIPPCLDSELTEFPLR VQLSPDLLATLPEPA SPGR GLFQVVSGGMVLQL QQGDQVWVEKDPK RSENVDSAEAVLKSE LNIQNAVNENQK ADGLAVIGVLMK TDLYFMPLAGSK NWETEITAQPDGGK * YGFQASNVTFIHGYI EKLGEAGIK CLKALQSLYTNRELF PK * 15 5 1 1 10. 2 4.7 0.526 1.4 4 -0.657 0.6 3 -0.765 0.5 9 Q08380. 1 15 6 1 1 10. 18 15.7 0 1.0 0 0 1.0 0 0 1.0 0 P01763. 1 15 7 1 1 10. 12 0 -0.445 0.7 3 1.795 3.4 7 1.804 3.4 9 Q8WZ4 2.4 64 associated protein 54 Galectin3-binding protein Ig heavy chain VIII region WEA Titin AAFGQGSGPIMLDEV QCTGTEASLADCK QVQLVDSGGGLVEP GGSLR NTADLKWTVPEKDG GSPITNYIVEK Table S7. Complete details of protein identification and label-free MS data for differentially expressed proteins in meningioma grade I (compared to healthy controls). Sl No . Nu m Spe ctr a Uni que Pep tide s Sco re 1 415 75 118 8.1 9 2 695 67 109 1.3 6 3 389 60 982 .32 Seq uenc e Cov erag e (%) 52.9 HC I (Set 1) HC I (Set 2) HC (Set 3) Aver age MG I (Set 1) MG I (Set 2) MG I (Set 3) Aver age Fol dcha nge 7.83 E+0 7 8.14 E+0 7 9.94 E+0 7 8636 6667 1.78 E+0 8 2.09 E+0 8 2.26 E+0 8 2043 3333 3 2.3 7 P010 24.2 74 3.23 E+0 8 3.52 E+0 8 4.20 E+0 8 3650 0000 0 6.00 E+0 8 7.29 E+0 8 7.63 E+0 8 6973 3333 3 1.9 1 P027 87.3 50 4.32 E+0 7 4.16 E+0 7 4.72 E+0 7 4400 0000 2.39 E+0 8 2.95 E+0 8 3.03 E+0 8 2790 0000 0 6.3 4 A5A6 I6.1 65 Protein Name Acces sion num ber RecName: Full=Complement C3: AltName: Full=C3 and PZPlike alpha-2-macroglobulin domain-containing protein 1: Contains: RecName: Full=Complement C3 beta chain: Contains: RecName: Full=Complement C3 alpha chain: Contains: RecName: Full=C3a anaphylatoxin: Contains: RecName: Full=Acylation stimulating protein: Short=ASP: AltName: Full=C3adesArg: Contains: RecName: Full=Complement C3b alpha' chain: Contains: RecName: Full=Complement C3c alpha' chain fragment 1: Contains: RecName: Full=Complement C3dg fragment: Contains: RecName: Full=Complement C3g fragment: Contains: RecName: Full=Complement C3d fragment: Contains: RecName: Full=Complement C3f fragment: Contains: RecName: Full=Complement C3c alpha' chain fragment 2: Flags: Precursor RecName: Full=Serotransferrin: Short=Transferrin: AltName: Full=Beta-1 metal-binding globulin: AltName: Full=Siderophilin: Flags: Precursor RecName: Full=Serotransferrin: Short=Transferrin: AltName: Full=Beta-1 metal-binding globulin: AltName: Full=Siderophilin: Flags: Precursor 4 269 36 600 .5 74.5 4.29 E+0 7 5.12 E+0 7 5.44 E+0 7 4950 0000 1.73 E+0 8 2.06 E+0 8 2.13 E+0 8 1973 3333 3 3.9 9 P010 23.3 5 294 33 501 .32 71.9 1.56 E+0 7 1.43 E+0 8 1.22 E+0 8 9353 3333 9.86 E+0 7 1.59 E+0 8 3.01 E+0 8 1862 0000 0 1.9 9 P026 47.1 6 237 30 448 .76 56.6 2.31 E+0 8 1.77 E+0 8 1.92 E+0 8 2000 0000 0 2.81 E+0 8 3.64 E+0 8 5.50 E+0 8 3983 3333 3 1.9 9 P0DJ G1.1 7 108 29 439 .72 26.8 1.61 E+0 7 1.07 E+0 7 1.07 E+0 7 1250 0000 4.63 E+0 7 4.79 E+0 7 3.98 E+0 7 4466 6667 3.5 7 P007 38.1 8 153 26 416 .22 48.7 48 1.04 E+0 8 7.00 E+0 7 1.04 E+0 8 9.71 E+0 7 P007 39.2 362 .25 7.00 E+0 7 6.07 E+0 7 6.9 8 21 1.47 E+0 7 5.70 E+0 7 9266 6667 159 1.12 E+0 7 7.18 E+0 7 1326 6667 9 1.39 E+0 7 5.23 E+0 7 7593 3333 1.2 6 P010 09.3 6036 6667 66 RecName: Full=Alpha-2macroglobulin: Short=Alpha-2-M: AltName: Full=C3 and PZPlike alpha-2-macroglobulin domain-containing protein 5: Flags: Precursor RecName: Full=Apolipoprotein A-I: Short=Apo-AI: Short=ApoA-I: AltName: Full=Apolipoprotein A1: Contains: RecName: Full=Truncated apolipoprotein A-I: AltName: Full=Apolipoprotein A-I(1242): Flags: Precursorgi|380876859|sp|G 3QY98.1|APOA1_GORGO RecName: Full=Apolipoprotein A-I: Short=Apo-AI: Short=ApoA-I: AltName: Full=Apolipoprotein A1: Contains: RecName: Full=Truncated apolipoprotein A-I: Flags: Precursorgi|385178604|sp|P 0DJG0.1|APOA1_PANTR RecName: Full=Apolipoprotein A-I: Short=Apo-AI: Short=ApoA-I: AltName: Full=Apolipoprotein A1: Contains: RecName: Full=Truncated apolipoprotein A-I: Flags: Precursor RecName: Full=Apolipoprotein A-I: Short=Apo-AI: Short=ApoA-I: AltName: Full=Apolipoprotein A1: Contains: RecName: Full=Truncated apolipoprotein A-I: Flags: Precursor RecName: Full=Haptoglobin: AltName: Full=Zonulin: Contains: RecName: Full=Haptoglobin alpha chain: Contains: RecName: Full=Haptoglobin beta chain: Flags: Precursor RecName: Full=Haptoglobin-related protein: Flags: Precursor RecName: Full=Alpha-1antitrypsin: AltName: Full=Alpha-1 protease inhibitor: AltName: Full=Alpha-1antiproteinase: AltName: Full=Serpin A1: Contains: RecName: Full=Short peptide from AAT: Short=SPAAT: Flags: Precursor 10 210 20 335 .36 59.7 8.36 E+0 7 9.92 E+0 7 9.28 E+0 7 9186 6667 1.67 E+0 8 1.97 E+0 8 2.82 E+0 8 2153 3333 3 2.3 4 Q5R CW5. 1 11 76 18 303 .55 30.6 1.81 E+0 7 8.96 E+0 6 1.02 E+0 7 1242 0000 3.17 E+0 7 3.27 E+0 7 3.18 E+0 7 3206 6667 2.5 8 P0C0 L4.1 12 117 18 302 .11 45.6 48.1 6.2 4 P018 71.3 RecName: Full=Ig mu chain C region 130 17 278 .6 57.9 P042 20.1 111 16 218 .42 32.7 1246 6666 7 8476 6667 2.4 1 15 2.0 3 P018 76.2 RecName: Full=Ig mu heavy chain disease protein: AltName: Full=BOT RecName: Full=Ig alpha-1 chain C region 16 57 15 197 .1 20.3 5666 666.7 1.3 5 P018 77.3 RecName: Full=Ig alpha-2 chain C region 17 109 14 243 .23 49.3 8.22 E+0 7 7.27 E+0 7 1.39 E+0 8 1.38 E+0 8 4.31 E+0 6 6.74 E+0 7 6720 0000 14 7.58 E+0 7 5.86 E+0 7 1.19 E+0 8 6.70 E+0 7 3.52 E+0 6 5.22 E+0 7 P027 68.2 271 .11 6.34 E+0 7 7.03 E+0 7 1.16 E+0 8 4.93 E+0 7 9.17 E+0 6 3.48 E+0 7 4.3 6 18 2.27 E+0 7 6.46 E+0 6 5.26 E+0 7 4.55 E+0 7 4.89 E+0 6 5.23 E+0 7 7380 0000 100 1.25 E+0 7 9.17 E+0 6 6.11 E+0 7 4.43 E+0 7 4.01 E+0 6 5.42 E+0 7 1693 3333 13 1.56 E+0 7 1.67 E+0 7 4.17 E+0 7 3.52 E+0 7 3.67 E+0 6 5.14 E+0 7 RecName: Full=Alpha-1antitrypsin: AltName: Full=Alpha-1 protease inhibitor: AltName: Full=Alpha-1antiproteinase: AltName: Full=Serpin A1: Flags: Precursor RecName: Full=Complement C4-A: AltName: Full=Acidic complement C4: AltName: Full=C3 and PZP-like alpha-2-macroglobulin domain-containing protein 2: Contains: RecName: Full=Complement C4 beta chain: Contains: RecName: Full=Complement C4-A alpha chain: Contains: RecName: Full=C4a anaphylatoxin: Contains: RecName: Full=C4b-A: Contains: RecName: Full=C4d-A: Contains: RecName: Full=Complement C4 gamma chain: Flags: Precursor RecName: Full=Serum albumin: Flags: Precursor 5146 6667 0.9 8 P004 50.1 18 82 14 232 .59 44.6 1.41 E+0 7 1.04 E+0 7 7.95 E+0 6 1081 6667 2.56 E+0 7 1.70 E+0 7 2.09 E+0 7 2116 6667 1.9 6 P027 90.2 19 142 13 207 .07 35.1 4.70 E+0 7 4.59 E+0 7 3.80 E+0 7 4363 3333 3.91 E+0 7 4.66 E+0 7 5.79 E+0 7 4786 6667 1.1 0 P027 74.1 RecName: Full=Ceruloplasmin: AltName: Full=Ferroxidase: Flags: Precursor RecName: Full=Hemopexin: AltName: Full=Beta-1B-glycoprotein: Flags: Precursor RecName: Full=Vitamin Dbinding protein: Short=DBP: Short=VDB: AltName: Full=Gc-globulin: AltName: Full=Groupspecific component: Flags: 1077 6667 5180 0000 4166 6667 4190 000 5263 3333 67 Precursor 20 123 12 225 .55 77.5 7.46 E+0 7 9.03 E+0 7 7.47 E+0 7 7986 6667 5.89 E+0 7 7.16 E+0 7 8.74 E+0 7 7263 3333 0.9 1 P086 03.4 21 83 12 194 .08 49.5 1.97 E+0 7 3.09 E+0 7 3.69 E+0 7 2916 6667 4.89 E+0 7 7.29 E+0 7 1.05 E+0 8 7560 0000 2.5 9 P010 08.1 22 31 12 133 .57 19.7 3.22 E+0 6 1.73 E+0 6 1.84 E+0 6 2263 333.3 5.88 E+0 6 8.03 E+0 6 7.99 E+0 6 7300 000 3.2 3 P042 17.4 23 30 11 168 .91 24.7 2.79 E+0 6 4.37 E+0 6 1.51 E+0 6 2890 000 1.44 E+0 7 1.91 E+0 7 2.06 E+0 7 1803 3333 6.2 4 P027 66.1 24 42 11 159 .52 4.5 6.53 E+0 5 1.31 E+0 6 1.08 E+0 6 1014 333.3 4.64 E+0 6 4.54 E+0 6 3.07 E+0 6 4083 333.3 4.0 3 P027 65.1 25 120 9 155 .12 92.4 3.41 E+0 7 4.64 E+0 7 4.60 E+0 7 4216 6667 6.89 E+0 7 9.70 E+0 7 8.10 E+0 7 8230 0000 1.9 5 Q9N2 D0.1 26 90 9 150 .39 74 6.31 E+0 7 7.04 E+0 7 4.29 E+0 7 5880 0000 1.74 E+0 8 1.90 E+0 8 1.77 E+0 8 1803 3333 3 3.0 7 P010 42.2 68 RecName: Full=Complement factor H: AltName: Full=H factor 1: Flags: Precursor RecName: Full=Antithrombin-III: Short=ATIII: AltName: Full=Serpin C1: Flags: Precursor RecName: Full=Alpha-1Bglycoprotein: AltName: Full=Alpha-1-B glycoprotein: Flags: Precursor RecName: Full=Transthyretin: AltName: Full=ATTR: AltName: Full=Prealbumin: AltName: Full=TBPA: Flags: Precursor RecName: Full=Alpha-2HS-glycoprotein: AltName: Full=Alpha-2-Z-globulin: AltName: Full=Ba-alpha-2glycoprotein: AltName: Full=Fetuin-A: Contains: RecName: Full=Alpha-2HS-glycoprotein chain A: Contains: RecName: Full=Alpha-2-HSglycoprotein chain B: Flags: Precursor RecName: Full=Alpha-2HS-glycoprotein: AltName: Full=Fetuin-A: Contains: RecName: Full=Alpha-2HS-glycoprotein chain A: Contains: RecName: Full=Alpha-2-HSglycoprotein chain B: Flags: Precursor RecName: Full=Kininogen1: AltName: Full=Alpha-2thiol proteinase inhibitor: AltName: Full=Fitzgerald factor: AltName: Full=High molecular weight kininogen: Short=HMWK: AltName: Full=Williams-FitzgeraldFlaujeac factor: Contains: RecName: Full=Kininogen1 heavy chain: Contains: RecName: Full=T-kinin: AltName: Full=Ile-SerBradykinin: Contains: RecName: Full=Bradykinin: AltName: Full=Kallidin I: Contains: RecName: Full=Lysyl-bradykinin: AltName: Full=Kallidin II: Contains: RecName: Full=Kininogen-1 light chain: Contains: RecName: Full=Low molecular weight growth-promoting factor: Flags: Precursor 27 51 7 128 .7 59.1 1.38 E+0 7 2.29 E+0 7 2.65 E+0 7 2106 6667 1.13 E+0 7 2.31 E+0 6 4.79 E+0 6 6133 333.3 0.2 9 P007 51.2 28 21 7 110 .17 17.5 8.27 E+0 5 3.09 E+0 6 4.38 E+0 6 2765 666.7 2.80 E+0 6 4.58 E+0 6 1.23 E+0 6 2870 000 1.0 4 P041 14.2 29 35 7 108 .84 37.3 16 1.89 E+0 7 1.77 E+0 6 2.06 E+0 7 3.66 E+0 6 P018 34.1 105 .07 1.44 E+0 7 1.05 E+0 6 1.0 9 7 1.34 E+0 7 2.39 E+0 5 1796 6667 17 1.94 E+0 7 8.54 E+0 5 1650 0000 30 1.67 E+0 7 4.77 E+0 5 2160 000 4.1 3 P026 52.1 5233 33.33 69 RecName: Full=Complement factor B: AltName: Full=C3/C5 convertase: AltName: Full=Glycine-rich beta glycoprotein: Short=GBG: AltName: Full=PBF2: AltName: Full=Properdin factor B: Contains: RecName: Full=Complement factor B Ba fragment: Contains: RecName: Full=Complement factor B Bb fragment: Flags: Precursor RecName: Full=Apolipoprotein B-100: Short=Apo B-100: Contains: RecName: Full=Apolipoprotein B-48: Short=Apo B-48: Flags: Precursor RecName: Full=Ig kappa chain C region RecName: Full=Apolipoprotein A-II: Short=Apo-AII: Short=ApoA-II: AltName: Full=Apolipoprotein A2: Contains: RecName: Full=Truncated apolipoprotein A-II: AltName: Full=Apolipoprotein A-II(176): Flags: Precursorgi|385178606|sp|P 0DJG2.1|APOA2_GORGO RecName: Full=Apolipoprotein A-II: Short=Apo-AII: Short=ApoA-II: AltName: Full=Apolipoprotein A2: Contains: RecName: Full=Truncated apolipoprotein A-II: Flags: Precursor 31 44 7 96. 93 16.7 1.14 E+0 7 6.57 E+0 6 8.33 E+0 6 8766 666.7 5.52 E+0 6 5.30 E+0 6 2.04 E+0 7 1040 6667 1.1 9 P688 71.2 32 18 7 82. 89 12.2 1.95 E+0 6 1.13 E+0 6 1.85 E+0 6 1643 333.3 1.17 E+0 6 8.22 E+0 5 1.50 E+0 6 1164 000 0.7 1 P040 04.1 33 16 7 81. 46 5.8 0.00 E+0 0 2.52 E+0 5 1.99 E+0 5 1503 33.33 3.74 E+0 6 4.67 E+0 6 4.54 E+0 6 4316 666.7 28. 71 P027 63.1 34 20 6 80. 1 22.8 1.32 E+0 6 0.00 E+0 0 1.11 E+0 6 8100 00 4.00 E+0 6 5.97 E+0 6 1.13 E+0 7 7090 000 8.7 5 P196 52.2 35 37 6 102 .86 31.3 9.30 E+0 6 1.21 E+0 7 7.83 E+0 6 9743 333.3 7.88 E+0 6 7.87 E+0 6 1.33 E+0 7 9683 333.3 0.9 9 P040 03.2 70 RecName: Full=Hemoglobin subunit beta: AltName: Full=Betaglobin: AltName: Full=Hemoglobin beta chain: Contains: RecName: Full=LVV-hemorphin7gi|56749857|sp|P68872.2|H BB_PANPA RecName: Full=Hemoglobin subunit beta: AltName: Full=Betaglobin: AltName: Full=Hemoglobin beta chaingi|56749858|sp|P68873 .2|HBB_PANTR RecName: Full=Hemoglobin subunit beta: AltName: Full=Betaglobin: AltName: Full=Hemoglobin beta chain RecName: Full=Vitronectin: Short=VN: AltName: Full=S-protein: AltName: Full=Serum-spreading factor: AltName: Full=V75: Contains: RecName: Full=Vitronectin V65 subunit: Contains: RecName: Full=Vitronectin V10 subunit: Contains: RecName: Full=Somatomedin-B: Flags: Precursor RecName: Full=Alpha-1acid glycoprotein 1: Short=AGP 1: AltName: Full=Orosomucoid-1: Short=OMD 1: Flags: Precursor RecName: Full=Alpha-1acid glycoprotein 2: Short=AGP 2: AltName: Full=Orosomucoid-2: Short=OMD 2: Flags: Precursor RecName: Full=C4bbinding protein alpha chain: Short=C4bp: AltName: Full=Proline-rich protein: Short=PRP: Flags: Precursor 36 84 6 100 .97 79.2 4.34 E+0 7 5.29 E+0 7 4.92 E+0 7 4850 0000 3.57 E+0 7 7.80 E+0 7 5.89 E+0 7 5753 3333 1.1 9 P109 09.1 37 20 6 98. 79 28.1 6.67 E+0 5 1.25 E+0 6 1.81 E+0 6 1242 333.3 3.32 E+0 6 2.23 E+0 6 2.76 E+0 6 2770 000 2.2 3 Q146 24.4 38 19 6 95. 94 14.6 6.59 E+0 5 6.93 E+0 5 7.32 E+0 5 6946 66.67 4.41 E+0 6 2.60 E+0 6 1.11 E+0 7 6036 666.7 8.6 9 P027 51.4 71 RecName: Full=Clusterin: AltName: Full=Agingassociated gene 4 protein: AltName: Full=Apolipoprotein J: Short=Apo-J: AltName: Full=Complement cytolysis inhibitor: Short=CLI: AltName: Full=Complementassociated protein SP-40,40: AltName: Full=Ku70binding protein 1: AltName: Full=NA1/NA2: AltName: Full=Testosterone-repressed prostate message 2: Short=TRPM-2: Contains: RecName: Full=Clusterin beta chain: AltName: Full=ApoJalpha: AltName: Full=Complement cytolysis inhibitor a chain: Contains: RecName: Full=Clusterin alpha chain: AltName: Full=ApoJbeta: AltName: Full=Complement cytolysis inhibitor b chain: Flags: Precursor RecName: Full=Inter-alphatrypsin inhibitor heavy chain H4: Short=ITI heavy chain H4: Short=ITI-HC4: Short=Inter-alpha-inhibitor heavy chain 4: AltName: Full=Inter-alpha-trypsin inhibitor family heavy chain-related protein: Short=IHRP: AltName: Full=Plasma kallikrein sensitive glycoprotein 120: Short=Gp120: Short=PK120: Contains: RecName: Full=70 kDa inter-alphatrypsin inhibitor heavy chain H4: Contains: RecName: Full=35 kDa inter-alphatrypsin inhibitor heavy chain H4: Flags: Precursor RecName: Full=Fibronectin: Short=FN: AltName: Full=Cold-insoluble globulin: Short=CIG: Contains: RecName: Full=Anastellin: Contains: RecName: Full=Ugl-Y1: Contains: RecName: Full=Ugl-Y2: Contains: RecName: Full=Ugl-Y3: Flags: Precursor 39 36 6 95. 42 19.2 4.68 E+0 6 1.98 E+0 6 1.60 E+0 7 7553 333.3 1.34 E+0 7 1.70 E+0 7 2.90 E+0 7 1980 0000 2.6 2 P027 49.3 40 10 6 87. 92 15.9 11.5 4.17 E+0 6 5.69 E+0 6 4.08 E+0 6 5.36 E+0 6 P0CG 05.1 75. 86 5.32 E+0 5 6.50 E+0 6 57. 78 6 1.52 E+0 5 0.00 E+0 0 2927 333.3 18 0.00 E+0 0 0.00 E+0 0 5066 6.667 41 0.00 E+0 0 6.22 E+0 5 5850 000 28. 22 P253 11.2 42 15 6 73. 75 14.8 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 3.44 E+0 6 4.06 E+0 6 3.06 E+0 6 3520 000 #DI V/0 ! P051 55.2 43 38 5 73. 61 49.2 1.68 E+0 7 1.48 E+0 7 1.43 E+0 7 1530 0000 2.75 E+0 6 1.73 E+0 6 3.49 E+0 6 2656 666.7 0.1 7 P007 34.2 44 23 4 71. 38 8 3.80 E+0 6 2.69 E+0 6 2.32 E+0 6 2936 666.7 1.25 E+0 7 1.17 E+0 7 1.42 E+0 7 1280 0000 4.3 6 P067 27.3 45 13 4 70. 74 10.4 4.55 E+0 5 3.38 E+0 6 1.62 E+0 6 1818 333.3 4.28 E+0 6 4.48 E+0 6 1.67 E+0 7 8486 666.7 4.6 7 P063 96.1 46 16 4 52. 19 11 1.48 E+0 6 8.83 E+0 5 3.41 E+0 5 9013 33.33 2.76 E+0 6 9.73 E+0 5 3.21 E+0 6 2314 333.3 2.5 7 P010 11.2 2073 33.33 72 RecName: Full=Beta-2glycoprotein 1: AltName: Full=APC inhibitor: AltName: Full=Activated protein C-binding protein: AltName: Full=Anticardiolipin cofactor: AltName: Full=Apolipoprotein H: Short=Apo-H: AltName: Full=Beta-2-glycoprotein I: Short=B2GPI: Short=Beta(2)GPI: Flags: Precursor RecName: Full=Ig lambda-2 chain C regions RecName: Full=Zinc-alpha2-glycoprotein: Short=Znalpha-2-GP: Short=Znalpha-2-glycoprotein: Flags: Precursor RecName: Full=Plasma protease C1 inhibitor: Short=C1 Inh: Short=C1Inh: AltName: Full=C1 esterase inhibitor: AltName: Full=C1-inhibiting factor: AltName: Full=Serpin G1: Flags: Precursor RecName: Full=Prothrombin: AltName: Full=Coagulation factor II: Contains: RecName: Full=Activation peptide fragment 1: Contains: RecName: Full=Activation peptide fragment 2: Contains: RecName: Full=Thrombin light chain: Contains: RecName: Full=Thrombin heavy chain: Flags: Precursor RecName: Full=Apolipoprotein A-IV: Short=Apo-AIV: Short=ApoA-IV: AltName: Full=Apolipoprotein A4: Flags: Precursor RecName: Full=Gelsolin: AltName: Full=AGEL: AltName: Full=Actindepolymerizing factor: Short=ADF: AltName: Full=Brevin: Flags: Precursor RecName: Full=Alpha-1antichymotrypsin: Short=ACT: AltName: Full=Cell growth-inhibiting gene 24/25 protein: AltName: Full=Serpin A3: Contains: RecName: Full=Alpha-1antichymotrypsin His-Pro- less: Flags: Precursor 47 3 3 20. 06 10.4 5.76 E+0 5 1.26 E+0 6 0.00 E+0 0 6120 00 1.73 E+0 8 2.06 E+0 8 0.00 E+0 0 1263 3333 3 206 .43 P699 05.2 48 15 3 59. 38 16.7 2.56 E+0 6 4.83 E+0 6 2.94 E+0 6 3443 333.3 3.98 E+0 6 2.78 E+0 6 7.89 E+0 5 2516 333.3 0.7 3 P198 23.2 49 9 3 43. 69 24 1.04 E+0 6 7.15 E+0 5 0.00 E+0 0 5850 00 1.47 E+0 6 5.62 E+0 5 4.97 E+0 5 8430 00 1.4 4 P007 47.2 50 5 3 42. 86 8.5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 6.26 E+0 5 2.51 E+0 6 2.28 E+0 5 1121 333.3 #DI V/0 ! P041 96.1 73 RecName: Full=Hemoglobin subunit alpha: AltName: Full=Alpha-globin: AltName: Full=Hemoglobin alpha chaingi|57013851|sp|P69906 .2|HBA_PANPA RecName: Full=Hemoglobin subunit alpha: AltName: Full=Alpha-globin: AltName: Full=Hemoglobin alpha chaingi|57013852|sp|P69907 .2|HBA_PANTR RecName: Full=Hemoglobin subunit alpha: AltName: Full=Alpha-globin: AltName: Full=Hemoglobin alpha chain RecName: Full=Inter-alphatrypsin inhibitor heavy chain H2: Short=ITI heavy chain H2: Short=ITI-HC2: Short=Inter-alpha-inhibitor heavy chain 2: AltName: Full=Inter-alpha-trypsin inhibitor complex component II: AltName: Full=Serum-derived hyaluronan-associated protein: Short=SHAP: Flags: Precursor RecName: Full=Plasminogen: Contains: RecName: Full=Plasmin heavy chain A: Contains: RecName: Full=Activation peptide: Contains: RecName: Full=Angiostatin: Contains: RecName: Full=Plasmin heavy chain A, short form: Contains: RecName: Full=Plasmin light chain B: Flags: Precursor RecName: Full=Histidinerich glycoprotein: AltName: Full=Histidine-proline-rich glycoprotein: Short=HPRG: Flags: Precursor 51 5 3 37. 97 18.2 9.41 E+0 5 0.00 E+0 0 5.99 E+0 5 5133 33.33 1.64 E+0 5 0.00 E+0 0 7.83 E+0 5 3156 66.67 0.6 1 P027 60.1 52 6 2 30. 2 22.8 25. 33 12.1 1.30 E+0 6 9.33 E+0 6 8.10 E+0 5 6.79 E+0 7 0.00 E+0 0 4.79 E+0 7 4171 0000 #DI V/0 ! 2.0 5 P181 36.2 2 0.00 E+0 0 2.16 E+0 7 7033 33.33 8 0.00 E+0 0 1.94 E+0 7 0 53 0.00 E+0 0 2.00 E+0 7 54 4 2 24. 91 1.9 24. 13 19.1 P016 25.2 2 19. 45 2.4 7300 0 0.6 5 P007 61.1 RecName: Full=Trypsin: Flags: Precursor 57 3 2 18. 25 5.7 1.28 E+0 5 4.65 E+0 6 2.19 E+0 5 1.26 E+0 6 8.2 0 2 1.38 E+0 5 2.36 E+0 5 0.00 E+0 0 0.00 E+0 0 3008 666.7 56 1.06 E+0 5 4.14 E+0 6 0.00 E+0 0 0.00 E+0 0 P015 91.4 2 6.09 E+0 4 5.92 E+0 5 0.00 E+0 0 5.20 E+0 5 6.1 1 10 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 4.47 E+0 5 1240 00 55 0.00 E+0 0 5.09 E+0 5 3.37 E+0 5 0.00 E+0 0 RecName: Full=Protein AMBP: Contains: RecName: Full=Alpha-1microglobulin: Short=Protein HC: AltName: Full=Alpha-1 microglycoprotein: AltName: Full=Complexforming glycoprotein heterogeneous in charge: Contains: RecName: Full=Inter-alpha-trypsin inhibitor light chain: Short=ITI-LC: AltName: Full=Bikunin: AltName: Full=EDC1: AltName: Full=HI-30: AltName: Full=Uronic-acid-rich protein: Contains: RecName: Full=Trypstatin: Flags: Precursor RecName: Full=Ig kappa chain V-III region HIC: Flags: Precursor RecName: Full=Afamin: AltName: Full=Alphaalbumin: Short=Alpha-Alb: Flags: Precursor RecName: Full=Immunoglobulin J chain: Flags: Precursor RecName: Full=Ig kappa chain V-IV region Len 4200 00 1.3 0 P010 31.4 58 3 2 15. 39 11.6 14. 31 0.6 3.17 E+0 4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 P017 66.1 2 0.00 E+0 0 3.84 E+0 5 0.0 2 2 7.15 E+0 5 0.00 E+0 0 1056 6.667 59 7.94 E+0 5 0.00 E+0 0 RecName: Full=Complement C5: AltName: Full=C3 and PZPlike alpha-2-macroglobulin domain-containing protein 4: Contains: RecName: Full=Complement C5 beta chain: Contains: RecName: Full=Complement C5 alpha chain: Contains: RecName: Full=C5a anaphylatoxin: Contains: RecName: Full=Complement C5 alpha' chain: Flags: Precursor RecName: Full=Ig heavy chain V-III region BRO 0 0.0 0 P051 56.2 2033 3333 2030 0 3670 00 1123 33.33 3223 33.33 5030 00 1280 00 74 P436 52.1 RecName: Full=Complement factor I: AltName: Full=C3B/C4B inactivator: Contains: RecName: Full=Complement factor I heavy chain: Contains: RecName: Full=Complement factor I light chain: Flags: Precursor 60 3 2 12. 62 7.1 0.00 E+0 0 0.00 E+0 0 8.39 E+0 5 2796 66.67 8.65 E+0 5 0.00 E+0 0 2.77 E+0 6 1211 666.7 4.3 3 O438 66.1 61 2 2 11. 96 4.7 0.00 E+0 0 0.00 E+0 0 5.69 E+0 5 1896 66.67 1.31 E+0 7 0.00 E+0 0 0.00 E+0 0 4366 666.7 23. 02 P042 07.2 62 1 1 8.7 7 2.2 5.3 8 2.6 0.00 E+0 0 2.34 E+0 6 3.16 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! #DI V/0 ! O750 37.2 1 0.00 E+0 0 0.00 E+0 0 1053 33.33 1 0.00 E+0 0 0.00 E+0 0 0 63 0.00 E+0 0 0.00 E+0 0 64 1 1 8.4 4 5.7 1 24. 82 38.8 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 9.69 E+0 7 4.00 E+0 4 9.14 E+0 6 0.00 E+0 0 0.00 E+0 0 1333 3.333 5 0.00 E+0 0 0.00 E+0 0 0 65 0.00 E+0 0 0.00 E+0 0 66 5 1 22. 18 39.3 1 18. 89 4.9 0.00 E+0 0 1.31 E+0 6 4.88 E+0 7 8.95 E+0 5 0.00 E+0 0 1.21 E+0 6 0.00 E+0 0 5.52 E+0 6 1626 6667 7 0.00 E+0 0 1.31 E+0 6 0 67 0.00 E+0 0 1.55 E+0 6 68 8 1 18. 77 7 1.74 E+0 6 7.49 E+0 5 3.80 E+0 6 2096 333.3 0.00 E+0 0 2.15 E+0 6 69 1 1 18. 4 3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 2.54 E+0 5 70 7 1 17. 46 4.7 1.94 E+0 6 1.05 E+0 6 2.37 E+0 6 1786 666.7 71 5 1 16. 14 1.2 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 72 1 1 15. 79 3.4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 7800 00 Q3S V33.1 #DI V/0 ! #DI V/0 ! Q86V V4.2 P517 77.1 2541 666.7 #DI V/0 ! 1.8 3 0.00 E+0 0 7166 66.67 0.3 4 P189 78.1 0.00 E+0 0 0.00 E+0 0 8466 6.667 #DI V/0 ! Q5R AD0. 1 2.53 E+0 5 0.00 E+0 0 0.00 E+0 0 8433 3.333 0.0 5 P074 15.1 0 6.10 E+0 5 7.10 E+0 5 4.22 E+0 5 5806 66.67 #DI V/0 ! P010 29.3 0 8.34 E+0 5 0.00 E+0 0 0.00 E+0 0 2780 00 #DI V/0 ! P191 34.4 0 1390 000 75 3534 6667 P320 81.1 P274 85.2 RecName: Full=CD5 antigen-like: AltName: Full=CT-2: AltName: Full=IgM-associated peptide: AltName: Full=SPalpha: Flags: Precursor RecName: Full=Ig kappa chain V-III region CLL: AltName: Full=Rheumatoid factor: Flags: Precursor RecName: Full=Kinesinlike protein KIF21B RecName: Full=Sulfite reductase [NADPH] hemoprotein betacomponent: Short=SiR-HP: Short=SiRHP RecName: Full=Ranbinding protein 3-like RecName: Full=Cold shock protein CspB: AltName: Full=Major cold shock protein RecName: Full=Cold shock protein CspD RecName: Full=Retinolbinding protein 4: AltName: Full=Plasma retinol-binding protein: Short=PRBP: Short=RBP: Flags: Precursor RecName: Full=Hemoglobin subunit alpha: AltName: Full=Alpha-globin: AltName: Full=Hemoglobin alpha chain RecName: Full=Complement component C7: Flags: Precursor RecName: Full=Hemoglobin subunit beta: AltName: Full=Betaglobin: AltName: Full=Hemoglobin beta chain RecName: Full=Complement C4-B: Contains: RecName: Full=Complement C4 beta chain: Contains: RecName: Full=Complement C4 alpha chain: Contains: RecName: Full=C4a anaphylatoxin: Contains: RecName: Full=Complement C4 gamma chain: Flags: Precursor RecName: Full=Serotransferrin: Short=Transferrin: AltName: Full=Beta-1 metal-binding globulin: AltName: Full=Siderophilin: Flags: Precursor 73 1 1 14. 86 2.7 3.71 E+0 5 0.00 E+0 0 0.00 E+0 0 1236 66.67 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q9C6 M5.1 74 2 1 14. 65 3.8 4.71 E+0 5 0.00 E+0 0 0.00 E+0 0 1570 00 1.45 E+0 6 0.00 E+0 0 0.00 E+0 0 4833 33.33 3.0 8 P086 97.3 75 1 1 12. 99 9.7 12. 92 7.2 1 12. 87 6.9 1.29 E+0 6 9.16 E+0 5 6.50 E+0 5 1198 666.7 2 0.00 E+0 0 2.68 E+0 6 0.00 E+0 0 #DI V/0 ! 6.8 9 77 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 P063 10.1 1 0.00 E+0 0 3.42 E+0 5 0.00 E+0 0 4300 00 6 0.00 E+0 0 1.80 E+0 5 0.00 E+0 0 0 76 0.00 E+0 0 0.00 E+0 0 1.01 E+0 5 2166 66.67 6.4 4 P355 42.2 78 9 1 12. 71 16.1 4.60 E+0 6 2.72 E+0 6 4.27 E+0 6 3863 333.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 P026 56.1 79 3 1 12. 39 0.9 0.00 E+0 0 1.73 E+0 6 1.83 E+0 6 1186 666.7 2.20 E+0 6 0.00 E+0 0 0.00 E+0 0 7333 33.33 0.6 2 O948 04.1 80 1 1 11. 67 11.6 11. 53 3.4 3.86 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! 0.0 0 P017 00.1 1 0.00 E+0 0 0.00 E+0 0 1286 66.67 1 0.00 E+0 0 0.00 E+0 0 0 81 0.00 E+0 0 8.07 E+0 6 A7HJ X5.1 82 4 1 11. 45 1.7 3.7 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 Q28H X4.1 11. 21 9.73 E+0 5 0.00 E+0 0 0.4 0 1 7.65 E+0 5 8.98 E+0 6 3243 33.33 7 7.39 E+0 5 4.87 E+0 6 8153 33.33 83 9.42 E+0 5 4.86 E+0 6 0 0.0 0 Q625 58.2 84 4 1 11. 07 0 3.69 E+0 5 4.61 E+0 5 6.08 E+0 5 4793 33.33 3.95 E+0 5 0.00 E+0 0 0.00 E+0 0 1316 66.67 0.2 7 Q9I7 U4.3 1740 00 3366 6.667 2690 000 6236 666.7 76 0 P807 48.1 RecName: Full=AP2/ERF and B3 domain-containing transcription repressor TEM1: AltName: Full=Protein TEMPRANILLO 1: AltName: Full=RAV1-like ethylene-responsive transcription factor TEM1 RecName: Full=Alpha-2antiplasmin: Short=Alpha-2AP: AltName: Full=Alpha2-plasmin inhibitor: Short=Alpha-2-PI: AltName: Full=Serpin F2: Flags: Precursor RecName: Full=Ig kappa chain V-II region RPMI 6410: Flags: Precursor RecName: Full=Ig lambda chain V-III region LOI RecName: Full=Serum amyloid A-4 protein: AltName: Full=Constitutively expressed serum amyloid A protein: Short=C-SAA: Flags: Precursor RecName: Full=Apolipoprotein C-III: Short=Apo-CIII: Short=ApoC-III: AltName: Full=Apolipoprotein C3: Flags: Precursor RecName: Full=Serine/threonineprotein kinase 10: AltName: Full=Lymphocyte-oriented kinase RecName: Full=Ig lambda chain V-I region HA RecName: Full=Valine-tRNA ligase: AltName: Full=Valyl-tRNA synthetase: Short=ValRS RecName: Full=Intraflagellar transport protein 57 homolog RecName: Full=Haptoglobin: AltName: Full=Zonulin: Contains: RecName: Full=Haptoglobin alpha chain: Contains: RecName: Full=Haptoglobin beta chain: Flags: Precursor RecName: Full=Titin: AltName: Full=D-Titin: AltName: Full=Kettin 85 1 1 10. 48 8.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 5.01 E+0 4 1670 0 #DI V/0 ! C1D RQ7. 1 86 1 1 10. 43 13.2 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 3.39 E+0 5 0.00 E+0 0 0.00 E+0 0 1130 00 #DI V/0 ! P590 93.1 87 1 1 10. 39 1.1 0.00 E+0 0 0.00 E+0 0 5.77 E+0 5 1923 33.33 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 A3P GJ0.1 88 1 1 10. 31 4.5 10. 07 0.5 0.0 0 1 10. 02 2.3 0 0.0 0 Q0P5 E6.1 91 1 1 10 13.8 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 Q0A BG0. 1 P286 85.1 90 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 4.14 E+0 5 0.0 0 1 0.00 E+0 0 3.11 E+0 6 0.00 E+0 0 0.00 E+0 0 0 3 1.65 E+0 6 1.45 E+0 6 2.57 E+0 6 0.00 E+0 0 5500 00 89 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1380 00 #DI V/0 ! B4U6 R8.1 92 1 1 9.8 9 0.6 0.00 E+0 0 4.71 E+0 5 0.00 E+0 0 1570 00 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q076 30.1 93 3 1 9.8 8 7.8 1.7 9.7 8 1 1 9.6 4 0.5 1.72 E+0 6 1.78 E+0 7 0.00 E+0 0 6816 666.7 95 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 B7VI S1.1 9.7 7 7.01 E+0 5 2.65 E+0 6 0.00 E+0 0 5.5 3 1 0.00 E+0 0 0.00 E+0 0 6.61 E+0 5 8070 00 4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1460 00 94 4.38 E+0 5 2.09 E+0 6 0.00 E+0 0 0 0.0 0 A6N DB9. 2 O515 68.1 96 1 1 9.6 2 2.9 0.00 E+0 0 0.00 E+0 0 5.99 E+0 5 1996 66.67 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q92B Q6.1 97 1 1 9.5 4 15.8 0.00 E+0 0 0.00 E+0 0 2.57 E+0 5 8566 6.667 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 B2HI J2.1 1520 000 8566 66.67 0 6966 66.67 2203 33.33 77 RecName: Full=Methylthioribose-1phosphate isomerase: Short=M1Pi: Short=MTR-1P isomerase: AltName: Full=S-methyl-5-thioribose1-phosphate isomerase RecName: Full=Uncharacterized protein C21orf99: AltName: Full=Cancer/testis antigen 85: Short=CT85 RecName: Full=DNAdirected RNA polymerase subunit beta': Short=RNAP subunit beta': AltName: Full=RNA polymerase subunit beta': AltName: Full=Transcriptase subunit beta' RecName: Full=50S ribosomal protein L6 RecName: Full=Contactin2: AltName: Full=Axonin-1: Flags: Precursor RecName: Full=Alpha-1syntrophin RecName: Full=Deoxyuridine 5'triphosphate nucleotidohydrolase: Short=dUTPase: AltName: Full=dUTP pyrophosphatase RecName: Full=RNA replication protein: AltName: Full=165 kDa protein: AltName: Full=ORF1 protein: Includes: RecName: Full=RNA-directed RNA polymerase: Includes: RecName: Full=Helicase RecName: Full=UPF0325 protein VS_2356 RecName: Full=Paralemmin-3: Flags: Precursor RecName: Full=Transcription-repaircoupling factor: Short=TRCF: AltName: Full=ATP-dependent helicase mfd RecName: Full=RNA polymerase sigma factor RpoD: AltName: Full=Sigma-43 RecName: Full=UracilDNA glycosylase: Short=UDG 98 3 1 9.5 4 3.7 6.91 E+0 5 0.00 E+0 0 0.00 E+0 0 2303 33.33 0.00 E+0 0 0.00 E+0 0 1.44 E+0 6 4800 00 2.0 8 P050 90.1 99 1 1 9.4 4 7.7 9.2 0.2 10 1 1 1 9.1 7 1.9 2.76 E+0 6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 3.83 E+0 6 2.47 E+0 6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! #DI V/0 ! #DI V/0 ! Q9HI R2.1 1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 9200 00 1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 10 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 10 2 5 1 9.1 5 14.9 4.00 E+0 6 0.00 E+0 0 0.00 E+0 0 1333 333.3 1.01 E+0 7 1.04 E+0 7 1.20 E+0 7 1083 3333 8.1 3 A9G UX6. 1 10 3 1 1 9.1 1 3.1 9.1 2.4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 4.94 E+0 6 4.10 E+0 3 2.18 E+0 6 #DI V/0 ! #DI V/0 ! Q5G H72.1 1 0.00 E+0 0 0.00 E+0 0 1366. 6667 3 0.00 E+0 0 0.00 E+0 0 0 10 4 0.00 E+0 0 0.00 E+0 0 10 5 2 1 9.0 3 1 4.35 E+0 5 0.00 E+0 0 6.52 E+0 5 3623 33.33 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 O433 45.1 10 6 1 1 8.8 6 3.6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 2.54 E+0 5 0.00 E+0 0 0.00 E+0 0 8466 6.667 #DI V/0 ! O433 74.2 10 7 5 1 8.8 1 14.2 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 7.58 E+0 6 7.33 E+0 6 2.09 E+0 6 5666 666.7 #DI V/0 ! Q8R BK6. 1 0 0 0 78 1276 666.7 8233 33.33 2373 333.3 Q54I X3.1 A1B4 95.1 P0AC P6.1 RecName: Full=Apolipoprotein D: Short=Apo-D: Short=ApoD: Flags: Precursor RecName: Full=50S ribosomal protein L2P RecName: Full=Probable polyketide synthase 26: Short=dipks26 RecName: Full=NADHquinone oxidoreductase subunit D: AltName: Full=NADH dehydrogenase I, subunit 4: AltName: Full=NADH dehydrogenase I, subunit D: AltName: Full=NADH-quinone oxidoreductase subunit 4: Short=NQO4: AltName: Full=NDH-1, subunit 4: AltName: Full=NDH-1, subunit D RecName: Full=NADHquinone oxidoreductase subunit B 1: AltName: Full=NADH dehydrogenase I subunit B 1: AltName: Full=NDH-1 subunit B 1 RecName: Full=XK-related protein 7 RecName: Full=HTH-type transcriptional regulator GntR: AltName: Full=Gluconate utilization system GNT-I transcriptional repressorgi|82581670|sp|P0 ACP5.1|GNTR_ECOLI RecName: Full=HTH-type transcriptional regulator GntR: AltName: Full=Gluconate utilization system GNT-I transcriptional repressor RecName: Full=Zinc finger protein 208: AltName: Full=Zinc finger protein 91like RecName: Full=Ras GTPase-activating protein 4: AltName: Full=Calciumpromoted Ras inactivator: AltName: Full=Ras p21 protein activator 4: AltName: Full=RasGAPactivating-like protein 2 RecName: Full=Phosphoribosylformyl glycinamidine synthase 1: AltName: Full=Phosphoribosylformyl glycinamidine synthase I: Short=FGAM synthase I 10 8 2 1 8.7 5 2.3 0.00 E+0 0 0.00 E+0 0 7.84 E+0 5 2613 33.33 6.23 E+0 5 0.00 E+0 0 0.00 E+0 0 2076 66.67 0.7 9 D2N T89.2 10 9 1 1 8.7 4 18.6 8.7 1 2.7 4.26 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 4.27 E+0 6 #DI V/0 ! #DI V/0 ! O070 06.1 1 0.00 E+0 0 0.00 E+0 0 1420 00 1 0.00 E+0 0 0.00 E+0 0 0 11 0 0.00 E+0 0 0.00 E+0 0 11 1 1 1 8.6 8 5.1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 4.61 E+0 6 1536 666.7 #DI V/0 ! Q8V V84.1 11 2 2 1 8.6 4 0.4 8.6 1 3 5 1 8.5 8 10.9 1.37 E+0 6 0.00 E+0 0 2.71 E+0 6 2.70 E+0 6 1.47 E+0 5 2.79 E+0 6 1833 333.3 #DI V/0 ! #DI V/0 ! 0.6 0 Q5RF M4.3 11 4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 Q8IY W2.3 1 0.00 E+0 0 0.00 E+0 0 2.72 E+0 6 1356 666.7 1 0.00 E+0 0 0.00 E+0 0 3.04 E+0 6 0 11 3 0.00 E+0 0 0.00 E+0 0 3.39 E+0 6 11 5 2 1 8.5 7 63 5.4 0.00 E+0 0 8.44 E+0 6 4.74 E+0 6 1.57 E+0 7 P015 42.2 8.5 7 0.00 E+0 0 0.00 E+0 0 0.5 3 1 8.98 E+0 6 7.96 E+0 6 1580 000 4 0.00 E+0 0 8.65 E+0 6 2993 333.3 11 6 0.00 E+0 0 0.00 E+0 0 8046 666.7 1.4 5 P622 58.1 11 7 3 1 8.5 6 1.2 5.30 E+0 5 0.00 E+0 0 0.00 E+0 0 1766 66.67 0.00 E+0 0 1.04 E+0 6 1.27 E+0 6 7700 00 4.3 6 O744 58.1 11 8 1 1 8.5 6 3.9 0.00 E+0 0.00 E+0 0.00 E+0 0 0.00 E+0 8.65 E+0 0.00 E+0 2883. 3333 #DI V/0 P465 57.1 0 0 3050 000 5536 666.7 79 1423 333.3 4900 0 Q5H PJ0.1 O578 18.1 RecName: Full=AAA ATPase forming ringshaped complexes: Short=ARC RecName: Full=Phenolic acid decarboxylase PadC: Short=PAD RecName: Full=Aconitate hydratase: Short=Aconitase: AltName: Full=Citrate hydro-lyase RecName: Full=60 kDa chaperonin: AltName: Full=GroEL protein: AltName: Full=Protein Cpn60 RecName: Full=Tetratricopeptide repeat protein 40 RecName: Full=Centromere/kinetochor e protein zw10 homolog RecName: Full=Uncharacterized HTHtype transcriptional regulator PH0045 RecName: Full=Crambin RecName: Full=14-3-3 protein epsilon: Short=14-33Egi|60391192|sp|P62259.1| 1433E_MOUSE RecName: Full=14-3-3 protein epsilon: Short=14-33Egi|61216932|sp|P62260.1| 1433E_RAT RecName: Full=14-3-3 protein epsilon: Short=14-3-3E: AltName: Full=Mitochondrial import stimulation factor L subunit: Short=MSF Lgi|71153779|sp|P62261.1|1 433E_BOVIN RecName: Full=14-3-3 protein epsilon: Short=14-33Egi|71153780|sp|P62262.1| 1433E_SHEEP RecName: Full=14-3-3 protein epsilon: Short=14-3-3E: AltName: Full=Protein kinase C inhibitor protein 1: Short=KCIP1gi|82197924|sp|Q5ZMT0.1 |1433E_CHICK RecName: Full=14-3-3 protein epsilon: Short=14-3-3E RecName: Full=Transcription factor tau subunit sfc4: AltName: Full=TFIIIC subunit sfc4: AltName: Full=Transcription factor C subunit 4 RecName: Full=Matrix nonpeptidase homolog 1: Flags: 0 0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0 3 0 3.57 E+0 5 2.20 E+0 6 1.35 E+0 5 0.00 E+0 0 7.60 E+0 4 0.00 E+0 0 1893 33.33 11 9 3 1 8.5 6 18.2 12 0 1 1 8.5 8.9 12 1 1 1 8.4 9 5.2 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 2.46 E+0 5 0.00 E+0 0 0.00 E+0 0 12 2 1 1 8.4 5 2.3 0.00 E+0 0 1.05 E+0 6 0.00 E+0 0 3500 00 0.00 E+0 0 0.00 E+0 0 12 3 1 1 8.4 4 0.5 1 1 8.4 1 0.9 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1220 00 12 4 3.66 E+0 5 0.00 E+0 0 0.00 E+0 0 5.13 E+0 6 12 5 1 1 8.3 8 2.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 12 6 6 1 8.3 1 34.8 1 1 8.3 1 1.3 12 8 3 1 8.2 9 0.3 12 9 2 1 8.2 8 8.7 13 0 1 1 8.2 1 3 2.64 E+0 6 0.00 E+0 0 0.00 E+0 0 6.84 E+0 5 0.00 E+0 0 6.14 E+0 6 0.00 E+0 0 0.00 E+0 0 7.74 E+0 5 0.00 E+0 0 3606 666.7 12 7 2.04 E+0 6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 13 1 2 1 8.1 7 0.3 1 1 8.1 5 2.9 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 13 2 0.00 E+0 0 2.40 E+0 5 0 0 0 0 4860 00 0 8000 0 ! Precursor #DI V/0 ! #DI V/0 ! Q9B YA7. 2 Q7TT U9.1 8200 0 #DI V/0 ! Q1M PL6.1 0.00 E+0 0 0 0.0 0 Q9UJ X2.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 P553 83.1 1710 000 #DI V/0 ! Q135 23.3 0.00 E+0 0 0.00 E+0 0 3.47 E+0 5 1156 66.67 #DI V/0 ! Q87R J4.1 7.16 E+0 6 1.51 E+0 6 7.81 E+0 5 0.00 E+0 0 0.00 E+0 0 6.70 E+0 6 0.00 E+0 0 1.35 E+0 6 0.00 E+0 0 0.00 E+0 0 6.70 E+0 6 0.00 E+0 0 2.26 E+0 6 0.00 E+0 0 4.67 E+0 4 6853 333.3 1.9 0 C4L2 G6.1 5033 33.33 #DI V/0 ! #DI V/0 ! 0.0 0 Q7T Q07.2 1556 6.667 #DI V/0 ! Q9Y4 D2.3 3.56 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 2.06 E+0 6 0.00 E+0 0 8053 33.33 #DI V/0 ! 0.0 0 P527 46.4 80 7333 33.33 1463 666.7 0 0 Q6B NL1. 2 P553 61.1 P427 68.4 RecName: Full=Putative proline dehydrogenase-like protein RecName: Full=Tryptophan--tRNA ligase: AltName: Full=Tryptophanyl-tRNA synthetase: Short=TrpRS RecName: Full=Aspartyl/glutamyltRNA(Asn/Gln) amidotransferase subunit B: Short=Asp/Glu-ADT subunit B RecName: Full=Cell division cycle protein 23 homolog: AltName: Full=Anaphase-promoting complex subunit 8: Short=APC8: AltName: Full=Cyclosome subunit 8 RecName: Full=Uncharacterized protein y4cA RecName: Full=Serine/threonineprotein kinase PRP4 homolog: AltName: Full=PRP4 kinase: AltName: Full=PRP4 premRNA-processing factor 4 homolog RecName: Full=DNA ligase: AltName: Full=Polydeoxyribonucleoti de synthase [NAD(+)] RecName: Full=Putative membrane protein insertion efficiency factor RecName: Full=DNA polymerase nu RecName: Full=Actin cytoskeleton-regulatory complex protein PAN1 RecName: Full=Probable transcriptional regulator SyrB RecName: Full=Sn1specific diacylglycerol lipase alpha: Short=DGLalpha: AltName: Full=Neural stem cellderived dendrite regulator RecName: Full=Zinc finger protein 142: AltName: Full=HA4654 RecName: Full=WiskottAldrich syndrome protein: Short=WASp 13 3 2 1 8.1 3 5.4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 1.98 E+0 5 4.93 E+0 5 0.00 E+0 0 2303 33.33 #DI V/0 ! Q96A E4.3 13 4 1 1 8.0 8 1.9 0.00 E+0 0 9.22 E+0 5 0.00 E+0 0 3073 33.33 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q6A E64.1 13 5 1 1 8.0 6 2.7 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 1.73 E+0 6 0.00 E+0 0 0.00 E+0 0 5766 66.67 #DI V/0 ! P102 44.1 13 6 1 1 8.0 5 0.4 0.00 E+0 0 3.06 E+0 5 0.00 E+0 0 1020 00 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q9B YW2. 3 13 7 4 1 8.0 4 2.5 0.00 E+0 0 3.56 E+0 5 4.49 E+0 5 2683 33.33 4.29 E+0 5 0.00 E+0 0 4.84 E+0 5 3043 33.33 1.1 3 P076 29.2 13 8 1 1 8.0 3 3.7 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 4.29 E+0 5 1430 00 #DI V/0 ! P359 13.2 13 9 1 1 8.0 2 3.7 8.0 1 3.1 14 1 2 1 7.9 6 1.1 3.77 E+0 4 8.06 E+0 5 5.75 E+0 5 0.00 E+0 0 1.10 E+0 6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! #DI V/0 ! 1.2 2 Q86U P0.1 1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1256 6.667 3 0.00 E+0 0 0.00 E+0 0 4.71 E+0 5 0 14 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 14 2 1 1 7.9 5 9.5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 1.87 E+0 6 6233 33.33 #DI V/0 ! O600 96.1 14 3 1 1 7.9 1 4.2 0.00 E+0 0.00 E+0 0.00 E+0 0 0.00 E+0 0.00 E+0 2.15 E+0 7166 66.67 #DI V/0 A3L Q86.2 0 1570 00 81 6353 33.33 1916 66.67 Q9N TN9. 1 P323 16.2 RecName: Full=Far upstream element-binding protein 1: Short=FBP: Short=FUSE-binding protein 1: AltName: Full=DNA helicase V: Short=hDH V RecName: Full=UDP-Nacetylglucosamine--Nacetylmuramyl(pentapeptide) pyrophosphorylundecaprenol Nacetylglucosamine transferase: AltName: Full=Undecaprenyl-PPMurNAc-pentapeptideUDPGlcNAc GlcNAc transferase RecName: Full=Mybrelated protein B: Short=BMyb: AltName: Full=Myblike protein 2 RecName: Full=Histonelysine N-methyltransferase SETD2: AltName: Full=HIF-1: AltName: Full=Huntingtin yeast partner B: AltName: Full=Huntingtin-interacting protein 1: Short=HIP-1: AltName: Full=Huntingtininteracting protein B: AltName: Full=Lysine Nmethyltransferase 3A: AltName: Full=SET domain-containing protein 2: Short=hSET2: AltName: Full=p231HBP RecName: Full=Serum amyloid P-component: AltName: Full=Female protein: Short=FP: AltName: Full=SAP(FP): Flags: Precursor RecName: Full=Rod cGMPspecific 3',5'-cyclic phosphodiesterase subunit beta: Short=GMP-PDE beta: Flags: Precursor RecName: Full=Cadherin24: Flags: Precursor RecName: Full=Semaphorin-4G: Flags: Precursor RecName: Full=Acetyl-CoA hydrolase: AltName: Full=Acetyl-CoA deacylase: Short=Acetyl-CoA acylase RecName: Full=37S ribosomal protein S35, mitochondrial: Flags: Precursor RecName: Full=Ribosome biogenesis protein YTM1 0 0 0 0 0 6 ! 14 4 1 1 7.8 9 1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 2.73 E+0 6 0.00 E+0 0 9100 00 #DI V/0 ! A4Y XQ9. 1 14 5 1 1 7.8 8 1.7 0.00 E+0 0 0.00 E+0 0 6.99 E+0 5 2330 00 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 A6N NM3. 3 14 6 1 1 7.8 6 1.6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 2.18 E+0 6 7266 66.67 #DI V/0 ! Q421 91.2 14 7 1 1 7.8 6 0.1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 3.41 E+0 6 0.00 E+0 0 0.00 E+0 0 1136 666.7 #DI V/0 ! Q8N F91.4 14 8 1 1 7.8 3 1.9 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 6.49 E+0 5 2163 33.33 #DI V/0 ! P024 57.3 14 9 1 1 7.8 1 5.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 4.28 E+0 5 1426 66.67 #DI V/0 ! B1YI T8.1 15 0 1 1 7.8 1 2.9 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 6.01 E+0 5 2003 33.33 #DI V/0 ! Q7V0 51.1 15 1 2 1 7.8 1 3 0.00 E+0 0 0.00 E+0 0 5.40 E+0 5 1800 00 0.00 E+0 0 0.00 E+0 0 2.02 E+0 6 6733 33.33 3.7 4 Q8IX Q6.2 15 2 1 1 7.8 4.9 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 5.31 E+0 5 0.00 E+0 0 0.00 E+0 0 1770 00 #DI V/0 ! Q68D Y1.2 82 RecName: Full=Glycine dehydrogenase [decarboxylating]: AltName: Full=Glycine cleavage system P-protein: AltName: Full=Glycine decarboxylase RecName: Full=RIMSbinding protein 3B: Short=RIM-BP3.B: AltName: Full=RIMSbinding protein 3.2: Short=RIM-BP3.2 RecName: Full=Mitochondrial inner membrane protein OXA1: AltName: Full=Oxidase assembly 1 protein: Short=AtOXA1: Flags: Precursor RecName: Full=Nesprin-1: AltName: Full=Enaptin: AltName: Full=Myocyte nuclear envelope protein 1: Short=Myne-1: AltName: Full=Nuclear envelope spectrin repeat protein 1: AltName: Full=Synaptic nuclear envelope protein 1: Short=Syne-1 RecName: Full=Collagen alpha-1(I) chain: AltName: Full=Alpha-1 type I collagen: Flags: Precursor RecName: Full=UDP-Nacetylmuramoylalanine--Dglutamate ligase: AltName: Full=D-glutamic acidadding enzyme: AltName: Full=UDP-Nacetylmuramoyl-L-alanylD-glutamate synthetase RecName: Full=2,3bisphosphoglycerateindependent phosphoglycerate mutase: Short=BPG-independent PGAM: Short=Phosphoglyceromuta se: Short=iPGM RecName: Full=Poly [ADPribose] polymerase 9: Short=PARP-9: AltName: Full=ADPribosyltransferase diphtheria toxin-like 9: Short=ARTD9: AltName: Full=B aggressive lymphoma protein RecName: Full=Zinc finger protein 626 15 3 1 1 7.7 4 0.8 0.00 E+0 0 0.00 E+0 0 1.91 E+0 6 6366 66.67 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 P225 16.1 15 4 1 1 7.7 3 5.1 0.00 E+0 0 7.06 E+0 6 0.00 E+0 0 2353 333.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 F2Z6 99.1 15 5 1 1 7.7 0.7 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 4.65 E+0 6 0.00 E+0 0 1550 000 #DI V/0 ! Q042 17.1 15 6 1 1 7.6 8 0.6 2.5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1.48 E+0 6 O603 03.4 7.6 0.00 E+0 0 0.00 E+0 0 0.0 0 1 5.57 E+0 5 0.00 E+0 0 0 1 0.00 E+0 0 0.00 E+0 0 1856 66.67 15 7 0.00 E+0 0 0.00 E+0 0 4933 33.33 #DI V/0 ! O141 33.3 15 8 1 1 7.5 9 1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 1.47 E+0 6 0.00 E+0 0 4900 00 #DI V/0 ! C3PF R3.1 15 9 1 1 7.5 8 8.1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 2.28 E+0 6 7600 00 #DI V/0 ! O001 70.2 16 0 1 1 7.5 7 0.6 8 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 C3PH 19.1 7.5 6 0.00 E+0 0 7.74 E+0 6 0.0 0 1 0.00 E+0 0 0.00 E+0 0 0 1 0.00 E+0 0 0.00 E+0 0 1006 66.67 16 1 3.02 E+0 5 0.00 E+0 0 2580 000 #DI V/0 ! Q7Z6 M1.1 16 2 1 1 7.5 2 3.6 5.7 0.00 E+0 0 2.38 E+0 6 0.00 E+0 0 0.00 E+0 0 Q8Y Q88.1 7.5 1 0.00 E+0 0 0.00 E+0 0 0.0 0 1 0.00 E+0 0 0.00 E+0 0 0 1 0.00 E+0 0 0.00 E+0 0 1990 00 16 3 5.97 E+0 5 0.00 E+0 0 7933 33.33 #DI V/0 ! Q039 24.5 0 0 0 83 RecName: Full=ATPdependent RNA helicase CHL1: AltName: Full=Chromosome loss protein 1: AltName: Full=Chromosome transmission fidelity protein 1 RecName: Full=External alternative NADHubiquinone oxidoreductase, mitochondrial: AltName: Full=External alternative NADH dehydrogenase: AltName: Full=NADH:ubiquinone reductase (nonelectrogenic): Flags: Precursor RecName: Full=Probable ATP-dependent RNA helicase DHR1: AltName: Full=DEAH box RNA helicase DHR1: AltName: Full=Extracellular mutant protein 16 RecName: Full=Uncharacterized protein KIAA0556 RecName: Full=ArginyltRNA--protein transferase 1: Short=Arginyltransferase 1: Short=R-transferase 1: AltName: Full=ArgininetRNA--protein transferase 1 RecName: Full=ATP synthase subunit alpha: AltName: Full=ATP synthase F1 sector subunit alpha: AltName: Full=FATPase subunit alpha RecName: Full=AH receptor-interacting protein: Short=AIP: AltName: Full=Aryl-hydrocarbon receptor-interacting protein: AltName: Full=HBV Xassociated protein 2: Short=XAP-2: AltName: Full=Immunophilin homolog ARA9 RecName: Full=Translation initiation factor IF-2 RecName: Full=Rab9 effector protein with kelch motifs: AltName: Full=40 kDa Rab9 effector protein: AltName: Full=p40 RecName: Full=Putative ABC transporter ATPbinding protein alr3946 RecName: Full=Zinc finger protein 117: AltName: Full=Provirus-linked krueppel: Short=h-PLK: AltName: Full=Zinc finger protein HPF9 16 4 1 1 7.5 1 1.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 1.01 E+0 6 0.00 E+0 0 3366 66.67 #DI V/0 ! Q0A C83.1 16 5 1 1 7.5 1 1.3 2.97 E+0 7 0.00 E+0 0 0.00 E+0 0 9900 000 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q5F WF4. 2 16 6 1 1 7.4 8 1.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 5.18 E+0 6 1726 666.7 #DI V/0 ! Q68C P9.2 16 7 1 1 7.4 8 4.4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 7.79 E+0 6 0.00 E+0 0 0.00 E+0 0 2596 666.7 #DI V/0 ! Q8J0 D2.1 16 8 1 1 7.4 1 0.7 0.00 E+0 0 0.00 E+0 0 1.07 E+0 6 3566 66.67 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q72K S4.1 16 9 1 1 7.4 7 7.3 8 6.5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1.41 E+0 6 0.00 E+0 0 #DI V/0 ! 0.0 0 Q2G3 S9.1 1 0.00 E+0 0 0.00 E+0 0 4700 00 1 0.00 E+0 0 0.00 E+0 0 0 17 0 0.00 E+0 0 6.02 E+0 6 17 1 1 1 7.3 7 11.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 1.73 E+0 5 0.00 E+0 0 5766 6.667 #DI V/0 ! Q137 90.2 17 2 1 1 7.3 7 2.9 7.3 7 6.7 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 3.25 E+0 6 0.00 E+0 0 0.00 E+0 0 0.0 0 1 6.82 E+0 5 0.00 E+0 0 0 1 0.00 E+0 0 0.00 E+0 0 2273 33.33 17 3 0.00 E+0 0 0.00 E+0 0 1083 333.3 #DI V/0 ! Q5X HC1. 1 Q9C BS7.1 2006 666.7 0 84 0 Q6V VB1. 2 RecName: Full=LPSassembly protein LptD: AltName: Full=Organic solvent tolerance protein: Flags: Precursor RecName: Full=DNA annealing helicase and endonuclease ZRANB3: AltName: Full=Annealing helicase 2: Short=AH2: AltName: Full=Zinc finger Ran-binding domaincontaining protein 3: Includes: RecName: Full=DNA annealing helicase ZRANB3: Includes: RecName: Full=Endonuclease ZRANB3 RecName: Full=AT-rich interactive domaincontaining protein 2: Short=ARID domaincontaining protein 2: AltName: Full=BRG1associated factor 200: Short=BAF200: AltName: Full=Zinc finger protein with activation potential: AltName: Full=Zipzap/p200 RecName: Full=Oligoxyloglucan reducing end-specific cellobiohydrolase: Short=OXG-RCBH: Flags: Precursor RecName: Full=Lon protease 1: AltName: Full=ATP-dependent protease La 1 RecName: Full=Trigger factor: Short=TF: AltName: Full=PPIase RecName: Full=E3 ubiquitin-protein ligase NHLRC1: AltName: Full=Malin: AltName: Full=NHL repeat-containing protein 1 RecName: Full=Apolipoprotein F: Short=Apo-F: AltName: Full=Lipid transfer inhibitor protein: Short=LTIP: Flags: Precursor RecName: Full=UPF0602 protein C4orf47 homolog RecName: Full=3-oxoacyl[acyl-carrier-protein] synthase 1: AltName: Full=Beta-ketoacyl-ACP synthase 1: Short=KAS 1 17 4 1 1 7.3 5 2.1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 5.97 E+0 6 0.00 E+0 0 0.00 E+0 0 17 5 1 1 7.3 3 2.6 17 6 1 1 7.3 2 0.8 17 7 2 1 7.3 3.8 17 8 1 1 7.2 9 17 9 1 1 18 0 1 18 1 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 5.77 E+0 6 0.00 E+0 0 1923 333.3 #DI V/0 ! 0.0 0 O945 25.1 1.00 E+0 6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 3333 33.33 0.00 E+0 0 1.16 E+0 6 0.00 E+0 0 1.03 E+0 6 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q8J1 G4.1 7300 00 #DI V/0 ! P398 20.3 6.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 4.92 E+0 5 0.00 E+0 0 1640 00 #DI V/0 ! O948 05.1 7.2 8 1.6 1 7.2 5 3.3 0.00 E+0 0 0.00 E+0 0 4.28 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1426 66.67 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1.97 E+0 6 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q98J M5.1 6566 66.67 #DI V/0 ! P432 84.1 3 1 7.2 5 6.7 5.50 E+0 5 0.00 E+0 0 7.35 E+0 5 4283 33.33 0.00 E+0 0 0.00 E+0 0 1.58 E+0 6 5266 66.67 1.2 3 Q6P4 M0.1 18 2 3 1 7.2 5 4 0.00 E+0 0 5.99 E+0 5 5.53 E+0 5 3840 00 1.09 E+0 6 0.00 E+0 0 0.00 E+0 0 3633 33.33 0.9 5 Q6P5 Z2.1 18 3 1 1 7.2 3 2.6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 2.75 E+0 5 0.00 E+0 0 0.00 E+0 0 9166 6.667 #DI V/0 ! O441 99.1 1990 000 0 0 85 0 P517 84.3 RecName: Full=Uncharacterized ribonuclease C609.01 RecName: Full=Ubiquitin carboxyl-terminal hydrolase 11: AltName: Full=Deubiquitinating enzyme 11: AltName: Full=Ubiquitin thioesterase 11: AltName: Full=Ubiquitin-specificprocessing protease 11 RecName: Full=Kinesinlike protein KIP1 RecName: Full=Glutamate 5-kinase 1: AltName: Full=Gamma-glutamyl kinase 1: Short=GK 1 RecName: Full=Actin-like protein 6B: AltName: Full=53 kDa BRG1associated factor B: AltName: Full=Actinrelated protein Baf53b: AltName: Full=ArpNalpha: AltName: Full=BRG1associated factor 53B: Short=BAF53B RecName: Full=DNA polymerase IV 2: Short=Pol IV 2 RecName: Full=Tryptophan synthase beta chain 2, chloroplastic: AltName: Full=Orange pericarp 2: Flags: Precursor RecName: Full=7dehydrocholesterol reductase: Short=7-DHC reductase: AltName: Full=Sterol Delta(7)reductase RecName: Full=Serine/threonineprotein kinase N3: AltName: Full=Protein kinase PKN-beta: AltName: Full=Protein-kinase Crelated kinase 3 RecName: Full=DNA repair protein rad-50 18 4 1 1 7.2 3 3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 3.13 E+0 6 0.00 E+0 0 0.00 E+0 0 1043 333.3 #DI V/0 ! Q029 28.1 18 5 3 1 7.2 2 2.1 0.00 E+0 0 6.31 E+0 6 0.00 E+0 0 2103 333.3 2.82 E+0 6 2.64 E+0 6 0.00 E+0 0 1820 000 0.8 7 Q96J 65.2 18 6 1 1 7.1 9 3.7 1 7.1 8 26.8 1 1 7.1 5 14.6 7.83 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 9.46 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! 0.0 0 18 8 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 2610 00 1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 18 7 0.00 E+0 0 1.45 E+0 5 0.00 E+0 0 Q7S HU8. 1 A0K EK5. 1 O958 16.1 18 9 1 1 7.1 5 10.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 1.71 E+0 6 19 0 1 1 7.1 5 2.7 0.00 E+0 0 1.29 E+0 6 0.00 E+0 0 4300 00 0.00 E+0 0 19 1 2 1 7.0 9 1 1.04 E+0 6 0.00 E+0 0 1.53 E+0 6 8566 66.67 19 2 1 1 7.0 7 2.5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 19 3 1 1 7.0 4 6.2 19 4 1 1 7.0 3 6 0.00 E+0 0 0.00 E+0 0 7.39 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 4833 3.333 0 0 3153 33.33 #DI V/0 ! 0.00 E+0 0 5700 00 #DI V/0 ! P529 54.2 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q0P4 K8.1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q3Y K19.1 0 0.00 E+0 0 1.92 E+0 4 0.00 E+0 0 6400 #DI V/0 ! E1BP 36.3 2463 33.33 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 2.62 E+0 6 0.00 E+0 0 0.00 E+0 0 0 0.0 0 P187 38.1 8733 33.33 #DI V/0 ! P0D D26.1 0 86 RecName: Full=Cytochrome P450 4A11: AltName: Full=20hydroxyeicosatetraenoic acid synthase: Short=20HETE synthase: AltName: Full=CYP4AII: AltName: Full=CYPIVA11: AltName: Full=Cytochrome P-450HKomega: AltName: Full=Cytochrome P450HLomega: AltName: Full=Fatty acid omegahydroxylase: AltName: Full=Lauric acid omegahydroxylase: Flags: Precursor RecName: Full=Multidrug resistance-associated protein 9: AltName: Full=ATPbinding cassette sub-family C member 12 RecName: Full=Probable dipeptidyl-aminopeptidase B: Short=DPAP B RecName: Full=Sulfurtransferase TusA homolog RecName: Full=BAG family molecular chaperone regulator 2: Short=BAG-2: AltName: Full=Bcl-2associated athanogene 2 RecName: Full=Transcription factor LBX1: AltName: Full=Ladybird homeobox protein homolog 1 RecName: Full=NFATC2interacting protein: AltName: Full=Nuclear factor of activated T-cells, cytoplasmic 2-interacting protein RecName: Full=Fanconi anemia group J protein homolog: Short=Protein FACJ: AltName: Full=ATPdependent RNA helicase BRIP1 RecName: Full=MMS19 nucleotide excision repair protein homolog: AltName: Full=MMS19-like protein RecName: Full=Gastrula zinc finger protein XlCGF9.1 RecName: Full=Probable dipeptidase Bgi|342165246|sp|P0DD27. 1|PEPDB_STRPQ RecName: Full=Probable dipeptidase B 19 5 1 1 7.0 1 3.5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 3.16 E+0 5 0.00 E+0 0 1053 33.33 #DI V/0 ! Q128 74.1 19 6 1 1 6.9 4 1.4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 1.03 E+0 7 3433 333.3 #DI V/0 ! B1LS 97.1 19 7 1 1 6.8 9 1.6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 2.82 E+0 5 0.00 E+0 0 9400 0 #DI V/0 ! Q9S D67.1 19 8 1 1 6.8 8 5.1 0.00 E+0 0 0.00 E+0 0 3.35 E+0 5 1116 66.67 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q03L V6.1 19 9 3 1 6.8 8 9.7 0.00 E+0 0 0.00 E+0 0 2.26 E+0 6 7533 33.33 7.31 E+0 4 8.86 E+0 6 0.00 E+0 0 2977 700 3.9 5 Q3Z WY8. 1 20 0 1 1 6.8 6 3.1 0.00 E+0 0 0.00 E+0 0 8.98 E+0 6 2993 333.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q128 51.2 20 1 1 1 6.8 2 0.5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 4.62 E+0 7 1540 0000 #DI V/0 ! Q96 M86. 2 20 2 1 1 6.8 2 17.1 1 6.7 8 2.8 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 2.56 E+0 6 0.00 E+0 0 0.00 E+0 0 1.05 E+0 4 0.00 E+0 0 3500 1 0.00 E+0 0 0.00 E+0 0 0 20 3 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! #DI V/0 ! Q9B TA0. 2 Q125 64.1 0 87 8533 33.33 RecName: Full=Splicing factor 3A subunit 3: AltName: Full=SF3a60: AltName: Full=Spliceosomeassociated protein 61: Short=SAP 61 RecName: Full=Sulfite reductase [NADPH] hemoprotein betacomponent: Short=SiR-HP: Short=SiRHP RecName: Full=ATPdependent zinc metalloprotease FTSH 7, chloroplastic: Short=AtFTSH7: Flags: Precursor RecName: Full=Glycine-tRNA ligase beta subunit: AltName: Full=GlycyltRNA synthetase beta subunit: Short=GlyRS RecName: Full=30S ribosomal protein S16gi|189044378|sp|A5FRP 7.1|RS16_DEHSB RecName: Full=30S ribosomal protein S16 RecName: Full=Mitogenactivated protein kinase kinase kinase kinase 2: AltName: Full=B lymphocyte serine/threonine-protein kinase: AltName: Full=Germinal center kinase: Short=GC kinase: AltName: Full=MAPK/ERK kinase kinase kinase 2: Short=MEK kinase kinase 2: Short=MEKKK 2: AltName: Full=Rab8interacting protein RecName: Full=Dynein heavy chain domaincontaining protein 1: AltName: Full=Dynein heavy chain domain 1-like protein: AltName: Full=Protein CCDC35 RecName: Full=Protein FAM167B RecName: Full=Chitin synthase A: AltName: Full=Chitin-UDP acetylglucosaminyl transferase A: AltName: Full=Class-I chitin synthase A 20 4 3 1 6.7 2 17.6 0.00 E+0 0 1.34 E+0 5 0.00 E+0 0 4466 6.667 0.00 E+0 0 5.78 E+0 5 0.00 E+0 0 1926 66.67 4.3 1 A1L1 C2.1 20 5 2 1 6.7 1 2.6 4.60 E+0 5 0.00 E+0 0 0.00 E+0 0 1533 33.33 2.93 E+0 6 0.00 E+0 0 0.00 E+0 0 9766 66.67 6.3 7 Q31V 62.1 20 6 1 1 6.7 1 6 1 6.7 2.8 0.00 E+0 0 0.00 E+0 0 1.05 E+0 5 6.26 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 3500 0 1 0.00 E+0 0 0.00 E+0 0 0 20 7 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! #DI V/0 ! Q9V AS7. 1 Q6T N15.1 20 8 1 1 6.6 9 4.9 3.84 E+0 5 0.00 E+0 0 0.00 E+0 0 1280 00 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 P598 28.1 20 9 1 1 6.6 7 0.6 6.6 1 1.1 0.00 E+0 0 0.00 E+0 0 1.84 E+0 6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! 0.0 0 Q8T DJ6.2 1 0.00 E+0 0 0.00 E+0 0 6133 33.33 1 0.00 E+0 0 0.00 E+0 0 0 21 0 0.00 E+0 0 1.07 E+0 6 21 1 1 1 6.5 9 6.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 3.81 E+0 5 1270 00 #DI V/0 ! Q8E VD0. 1 21 2 3 1 6.5 6 6.7 0.00 E+0 0 1.92 E+0 6 1.95 E+0 6 1290 000 0.00 E+0 0 0.00 E+0 0 2.20 E+0 6 7333 33.33 0.5 7 A3C T71.1 21 3 1 1 6.5 6 1.6 6.5 6 1.2 5.10 E+0 6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! 0.0 0 Q7Z3 92.2 1 0.00 E+0 0 5.65 E+0 5 1700 000 2 0.00 E+0 0 0.00 E+0 0 0 21 4 0.00 E+0 0 3.51 E+0 5 Q9C0 99.2 21 5 1 1 6.5 5 7.5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 7.18 E+0 5 2393 33.33 #DI V/0 ! C1A UB5. 1 0 3566 66.67 3053 33.33 0 88 2086 66.67 0 0 Q562 F6.2 RecName: Full=Mitochondrial cardiolipin hydrolase: AltName: Full=Choline phosphatase 6: AltName: Full=Mitochondrial phospholipase: Short=MitoPLD: AltName: Full=Phosphatidylcholinehydrolyzing phospholipase D6: AltName: Full=Phospholipase D6: Short=PLD 6 RecName: Full=Glycine-tRNA ligase beta subunit: AltName: Full=GlycyltRNA synthetase beta subunit: Short=GlyRS RecName: Full=Innexin inx3: Short=Innexin-3 RecName: Full=Interferoninduced GTP-binding protein Mx: AltName: Full=Interferon-inducible Mx protein RecName: Full=Poly(betaD-mannuronate) C5 epimerase: Flags: Precursor RecName: Full=DmX-like protein 2: AltName: Full=Rabconnectin-3 RecName: Full=Shugoshinlike 2: AltName: Full=Shugoshin-2: Short=Sgo2: AltName: Full=Tripin RecName: Full=Glycerol kinase: AltName: Full=ATP:glycerol 3phosphotransferase: AltName: Full=Glycerokinase: Short=GK RecName: Full=Probable endonuclease 4: AltName: Full=Endodeoxyribonucleas e IV: AltName: Full=Endonuclease IV RecName: Full=Trafficking protein particle complex subunit 11 RecName: Full=Leucinerich repeat and coiled-coil domain-containing protein 1: AltName: Full=Centrosomal leucinerich repeat and coiled-coil domain-containing protein RecName: Full=Probable cytosol aminopeptidase: AltName: Full=Leucine aminopeptidase: Short=LAP: AltName: Full=Leucyl aminopeptidase 21 6 1 1 6.5 4 0.5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1.91 E+0 6 5.80 E+0 5 6366 66.67 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 O751 28.2 RecName: Full=Protein cordon-bleu 21 7 1 1 6.5 4 0.8 0 0.0 0 Q8N DV3. 2 3666 66.67 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q9SI V5.3 6.04 E+0 5 0.00 E+0 0 2013 33.33 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1.79 E+0 6 0.00 E+0 0 0.00 E+0 0 0 0.0 0 P507 68.1 RecName: Full=Structural maintenance of chromosomes protein 1B: Short=SMC protein 1B: Short=SMC-1-beta: Short=SMC-1B RecName: Full=Zinc finger CCCH domain-containing protein 19: Short=AtC3H19: AltName: Full=Protein Needed for RDR2independent DNA methylation RecName: Full=Regulatory protein E2 21 8 1 1 6.5 4 0.3 1.10 E+0 6 0.00 E+0 0 0.00 E+0 0 21 9 1 1 6.5 3 4.5 1 6.5 1.2 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 22 0 1 5966 66.67 #DI V/0 ! Q5JS H3.1 22 1 1 1 6.4 5 1.1 3.84 E+0 5 0.00 E+0 0 0.00 E+0 0 1280 00 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 P891 05.3 22 2 1 1 6.4 4 0.4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 2.12 E+0 5 0.00 E+0 0 0.00 E+0 0 7066 6.667 #DI V/0 ! Q75J N1.1 22 3 1 1 6.4 3 1.7 0.00 E+0 0 4.17 E+0 6 0.00 E+0 0 1390 000 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q141 88.2 22 4 2 1 6.4 2 5.4 4.66 E+0 5 8.45 E+0 5 0.00 E+0 0 4370 00 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 A1R Y72.1 22 5 1 1 6.4 2 1.4 4.23 E+0 5 0.00 E+0 0 0.00 E+0 0 1410 00 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q8B TM8. 5 22 6 1 1 6.4 2 1.7 0.8 0.00 E+0 0 8.51 E+0 6 0.00 E+0 0 0.00 E+0 0 Q7U ZL6.1 6.4 2 0.00 E+0 0 0.00 E+0 0 0.0 0 1 0.00 E+0 0 0.00 E+0 0 0 1 0.00 E+0 0 0.00 E+0 0 1603 33.33 22 7 4.81 E+0 5 0.00 E+0 0 2836 666.7 #DI V/0 ! Q9U PN4. 3 22 8 1 1 6.4 1 3.9 0.00 E+0 0.00 E+0 0.00 E+0 0 3.15 E+0 0.00 E+0 0.00 E+0 1050 000 #DI V/0 C4K4 F9.1 1933 33.33 0 0 89 RecName: Full=WD repeatcontaining protein 44: AltName: Full=Rabphilin11 RecName: Full=RNA polymerase-associated protein CTR9: AltName: Full=Centromere-binding factor 1-dependent protein 1: AltName: Full=Cln threerequiring protein 9 RecName: Full=Probable serine/threonine-protein kinase ifkC: AltName: Full=Initiation factor kinase C RecName: Full=Transcription factor Dp-2: AltName: Full=E2F dimerization partner 2 RecName: Full=DNA ligase 2: AltName: Full=Polydeoxyribonucleoti de synthase [ATP] 2 RecName: Full=Filamin-A: Short=FLN-A: AltName: Full=Actin-binding protein 280: Short=ABP-280: AltName: Full=Alphafilamin: AltName: Full=Endothelial actinbinding protein: AltName: Full=Filamin-1: AltName: Full=Non-muscle filamin RecName: Full=DNA mismatch repair protein MutS RecName: Full=5azacytidine-induced protein 1: AltName: Full=Centrosomal protein of 131 kDa: Short=Cep131: AltName: Full=Preacrosome localization protein 1 RecName: Full=Elongation factor G: Short=EF-G 0 0 0 6 0 0 ! 22 9 2 1 6.3 9 17.9 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 1.45 E+0 6 1.37 E+0 6 9400 00 #DI V/0 ! O065 89.1 23 0 3 1 6.3 9 0.8 6.3 8 1.8 0.0 0 1 6.3 8 5.2 5.18 E+0 5 0.00 E+0 0 1.02 E+0 5 0 1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 Q7V NT4. 1 O760 83.1 23 2 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.6 6 1 7.88 E+0 5 0.00 E+0 0 0.00 E+0 0 1726 66.67 2 0.00 E+0 0 1.12 E+0 7 0.00 E+0 0 2626 66.67 23 1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 3400 0 #DI V/0 ! Q54S A1.1 23 3 1 1 6.3 7 1.1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 1.51 E+0 6 0.00 E+0 0 0.00 E+0 0 5033 33.33 #DI V/0 ! P611 34.1 23 4 1 1 6.3 7 1.7 7.7 0.00 E+0 0 6.95 E+0 5 0.00 E+0 0 0.00 E+0 0 Q752 Q3.2 6.3 3 0.00 E+0 0 0.00 E+0 0 0.0 0 1 0.00 E+0 0 0.00 E+0 0 0 1 0.00 E+0 0 0.00 E+0 0 1276 66.67 23 5 3.83 E+0 5 0.00 E+0 0 2316 66.67 #DI V/0 ! Q003 25.2 23 6 1 1 6.3 2 3.8 1.59 E+0 5 0.00 E+0 0 0.00 E+0 0 5300 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q9K UR9. 2 23 7 1 1 6.3 1 4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 7.73 E+0 5 0.00 E+0 0 0.00 E+0 0 2576 66.67 #DI V/0 ! B3L2 G0.1 23 8 1 1 6.3 1 1.8 6.79 E+0 6 0.00 E+0 0 0.00 E+0 0 2263 333.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q96Q 42.2 3733 333.3 0 0 90 RecName: Full=Imidazole glycerol phosphate synthase subunit HisH: AltName: Full=IGP synthase glutamine amidotransferase subunit: AltName: Full=IGP synthase subunit HisH: AltName: Full=ImGP synthase subunit HisH: Short=IGPS subunit HisH RecName: Full=Electron transport complex protein RnfC RecName: Full=High affinity cGMP-specific 3',5'cyclic phosphodiesterase 9A RecName: Full=Phospholipase D Z: AltName: Full=Phosphatase D3: Short=PLD 3: Flags: Precursor RecName: Full=Complement component C6: Flags: Precursor RecName: Full=Enhancer of polycomb-like protein 1 RecName: Full=Phosphate carrier protein, mitochondrial: AltName: Full=Phosphate transport protein: Short=PTP: AltName: Full=Solute carrier family 25 member 3: Flags: Precursor RecName: Full=LPSassembly protein LptD: AltName: Full=Organic solvent tolerance protein: Flags: Precursor RecName: Full=tRNA (guanine(37)-N1)methyltransferase: AltName: Full=M1Gmethyltransferase: AltName: Full=tRNA [GM37] methyltransferase: AltName: Full=tRNA methyltransferase 5 homolog RecName: Full=Alsin: AltName: Full=Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 6 protein: AltName: Full=Amyotrophic lateral sclerosis 2 protein 23 9 2 1 6.2 9 4.2 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 2.09 E+0 6 0.00 E+0 0 1.94 E+0 6 1343 333.3 #DI V/0 ! Q6X PS3.2 24 0 1 1 6.2 9 4.5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 2.08 E+0 5 6933 3.333 #DI V/0 ! Q8N FI3.1 24 1 1 1 6.2 4 6.7 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 4.48 E+0 5 0.00 E+0 0 0.00 E+0 0 1493 33.33 #DI V/0 ! Q498 42.2 24 2 1 1 6.2 4 2.3 0.00 E+0 0 2.81 E+0 4 0.00 E+0 0 9366. 6667 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q929 95.2 24 3 1 1 6.2 2.1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 8.38 E+0 5 0.00 E+0 0 0.00 E+0 0 2793 33.33 #DI V/0 ! Q68D V7.1 24 4 1 1 6.2 3.7 1 6.1 9 2 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 5.83 E+0 5 7.34 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 2446 66.67 1 0.00 E+0 0 0.00 E+0 0 0 24 5 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! #DI V/0 ! Q9Z DC9. 1 Q6Z V73.2 24 6 1 1 6.1 6 1.5 2.2 0.00 E+0 0 3.83 E+0 5 0.00 E+0 0 0.00 E+0 0 O821 39.1 6.1 5 0.00 E+0 0 0.00 E+0 0 0.0 0 1 0.00 E+0 0 0.00 E+0 0 0 1 3.72 E+0 5 0.00 E+0 0 1240 00 24 7 0.00 E+0 0 0.00 E+0 0 1276 66.67 #DI V/0 ! Q025 12.2 24 8 1 1 6.1 3 3.8 0.00 E+0 0 4.49 E+0 5 0.00 E+0 0 1496 66.67 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 B4U9 K9.1 24 9 1 1 6.1 13.6 0.00 E+0 0 2.17 E+0 5 0.00 E+0 0 7233 3.333 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 A4X TF3.1 0 0 91 1943 33.33 RecName: Full=Phosphatidylinositol 3,4,5-trisphosphate 3phosphatase TPTE2: AltName: Full=Lipid phosphatase TPIP: AltName: Full=TPTE and PTEN homologous inositol lipid phosphatase RecName: Full=Cytosolic endo-beta-Nacetylglucosaminidase: Short=ENGase RecName: Full=(Dimethylallyl)adenosi ne tRNA methylthiotransferase MiaB: AltName: Full=tRNAi(6)A37 methylthiotransferase RecName: Full=Ubiquitin carboxyl-terminal hydrolase 13: AltName: Full=Deubiquitinating enzyme 13: AltName: Full=Isopeptidase T-3: Short=ISOT-3: AltName: Full=Ubiquitin thioesterase 13: AltName: Full=Ubiquitin-specificprocessing protease 13 RecName: Full=E3 ubiquitin-protein ligase RNF43: AltName: Full=RING finger protein 43: Flags: Precursor RecName: Full=Putative carboxypeptidase RP402 RecName: Full=FYVE, RhoGEF and PH domaincontaining protein 6: AltName: Full=Zinc finger FYVE domain-containing protein 24 RecName: Full=Cycloartenol Synthase RecName: Full=Replication protein E1: AltName: Full=ATP-dependent helicase E1 RecName: Full=Adenylosuccinate synthetase: Short=AMPSase: Short=AdSS: AltName: Full=IMP--aspartate ligase RecName: Full=Integration host factor subunit beta: Short=IHF-beta 25 0 1 1 6.1 0.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 4.73 E+0 5 0.00 E+0 0 0.00 E+0 0 1576 66.67 #DI V/0 ! O755 92.3 25 1 1 1 6.0 7 1.6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 6.62 E+0 5 0.00 E+0 0 0.00 E+0 0 2206 66.67 #DI V/0 ! Q9Y2 H2.3 25 2 1 1 6.0 6 1.4 6.0 5 4.6 0.00 E+0 0 4.47 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.0 0 1 0.00 E+0 0 0.00 E+0 0 0 1 0.00 E+0 0 0.00 E+0 0 2356 66.67 25 3 7.07 E+0 5 0.00 E+0 0 1490 00 #DI V/0 ! Q9L VR3. 1 P352 49.2 25 4 1 1 6 2.5 2.1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 8.30 E+0 5 Q7U A23.1 5.9 9 0.00 E+0 0 0.00 E+0 0 0.0 0 1 6.11 E+0 5 0.00 E+0 0 0 1 0.00 E+0 0 0.00 E+0 0 2036 66.67 25 5 0.00 E+0 0 0.00 E+0 0 2766 66.67 #DI V/0 ! O150 67.4 25 6 1 1 5.9 8 1.7 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 1.13 E+0 6 0.00 E+0 0 0.00 E+0 0 3766 66.67 #DI V/0 ! Q8W TR7. 1 25 7 1 1 5.9 7 5.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 3.81 E+0 5 1270 00 #DI V/0 ! Q4G1 76.3 25 8 1 1 5.9 7 1.3 1 5.9 7 2.4 0.00 E+0 0 0.00 E+0 0 3.62 E+0 5 4.27 E+0 6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1206 66.67 1 0.00 E+0 0 0.00 E+0 0 0 25 9 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! #DI V/0 ! Q5V XU9. 1 Q6U XZ4. 1 0 0 0 92 1423 333.3 RecName: Full=Probable E3 ubiquitin-protein ligase MYCBP2: AltName: Full=Myc-binding protein 2: AltName: Full=Pam/highwire/rpm-1 protein: AltName: Full=Protein associated with Myc RecName: Full=Phosphatidylinositide phosphatase SAC2: AltName: Full=Inositol polyphosphate 5phosphatase F: AltName: Full=Sac domain-containing inositol phosphatase 2: AltName: Full=Sac domaincontaining phosphoinositide 5-phosphatase 2: Short=hSAC2 RecName: Full=UPF0496 protein At5g66670 RecName: Full=Replication factor C subunit 4: AltName: Full=Activator 1 37 kDa subunit: Short=A1 37 kDa subunit: AltName: Full=Activator 1 subunit 4: AltName: Full=Replication factor C 37 kDa subunit: Short=RF-C 37 kDa subunit: Short=RFC37 RecName: Full=DNA mismatch repair protein MutS RecName: Full=Phosphoribosylformyl glycinamidine synthase: Short=FGAM synthase: Short=FGAMS: AltName: Full=Formylglycinamide ribotide amidotransferase: Short=FGARAT: AltName: Full=Formylglycinamide ribotide synthetase RecName: Full=Zinc finger protein 473: AltName: Full=Zinc finger protein 100 homolog: Short=Zfp-100 RecName: Full=Acyl-CoA synthetase family member 3, mitochondrial: Flags: Precursor RecName: Full=Uncharacterized protein C9orf84 RecName: Full=Netrin receptor UNC5D: AltName: Full=Protein unc-5 homolog 4: AltName: Full=Protein unc-5 homolog D: Flags: Precursor 26 0 1 1 5.9 6 1.8 8.50 E+0 5 0.00 E+0 0 0.00 E+0 0 2833 33.33 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 P263 58.2 26 1 1 1 5.9 5 9.9 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 2.77 E+0 6 9233 33.33 #DI V/0 ! O437 09.2 26 2 1 1 5.9 5 1.5 8.5 0.0 0 P367 81.1 1 1 5.9 4 10.7 0.00 E+0 0 0.00 E+0 0 3.55 E+0 5 0 26 4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 P323 86.2 5.9 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.0 0 1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1076 666.7 26 3 3.23 E+0 6 1.59 E+0 5 0.00 E+0 0 1183 33.33 #DI V/0 ! P091 24.2 26 5 1 1 5.9 2 3.8 1 5.9 2 12.6 26 7 1 1 5.9 2.6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 3.54 E+0 5 3.83 E+0 5 1.70 E+0 6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1180 00 1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 26 6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! #DI V/0 ! #DI V/0 ! Q0W VN5. 2 Q1M PQ7. 1 Q5D RC2. 1 26 8 1 1 5.8 8 20.8 5.8 7 3.2 1 1 5.8 6 4.1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! 0.0 0 27 0 4.35 E+0 4 0.00 E+0 0 0.00 E+0 0 Q9BP H3.1 1 0.00 E+0 0 0.00 E+0 0 6.30 E+0 5 1450 0 1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 26 9 0.00 E+0 0 1.52 E+0 7 0.00 E+0 0 0 0.0 0 27 1 1 1 5.8 5 4.9 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1.79 E+0 5 5966 6.667 #DI V/0 ! 5300 0 0 0 0 5066 666.7 2100 00 0 93 1276 66.67 5666 66.67 0 Q6Z W76. 1 Q8K9 B2.1 O947 76.1 RecName: Full=DNA (cytosine-5)methyltransferase 1: Short=Dnmt1: AltName: Full=CXXC-type zinc finger protein 9: AltName: Full=DNA methyltransferase HsaI: Short=DNA MTase HsaI: Short=M.HsaI: AltName: Full=MCMT RecName: Full=Uncharacterized methyltransferase WBSCR22: AltName: Full=Williams-Beuren syndrome chromosomal region 22 protein RecName: Full=ATPdependent bile acid permease RecName: Full=Regulatory protein E2 RecName: Full=Glyceraldehyde-3phosphate dehydrogenase 1: AltName: Full=NADdependent glyceraldehyde3-phosphate dehydrogenase: Short=GAPDH RecName: Full=Cellulose synthase-like protein G3: Short=AtCslG3 RecName: Full=30S ribosomal protein S3 RecName: Full=Protocadherin gammaA1: Short=PCDH-gammaA1: Flags: Precursor RecName: Full=Conotoxin ArMLKM-01: Flags: Precursor RecName: Full=Ankyrin repeat and SAM domaincontaining protein 3 RecName: Full=Fructosebisphosphate aldolase class 2: Short=FBP aldolase: Short=FBPA: AltName: Full=Fructose-1,6bisphosphate aldolase: AltName: Full=Fructosebisphosphate aldolase class II RecName: Full=Metastasisassociated protein MTA2: AltName: Full=Metastasisassociated 1-like 1: Short=MTA1-L1 protein: AltName: Full=p53 target protein in deacetylase complex 27 2 1 1 5.8 5 3.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 4.83 E+0 5 0.00 E+0 0 0.00 E+0 0 1610 00 #DI V/0 ! Q095 01.1 27 3 1 1 5.8 5 1.1 0.00 E+0 0 4.72 E+0 5 0.00 E+0 0 1573 33.33 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q146 78.3 27 4 1 1 5.8 4 3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 5.91 E+0 5 1970 00 #DI V/0 ! P307 60.2 27 5 1 1 5.8 2 8.5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 3.55 E+0 5 1183 33.33 #DI V/0 ! C3K3 34.1 27 6 1 1 5.8 2 6.6 0.7 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 Q9L V41.1 5.7 6 0.00 E+0 0 0.00 E+0 0 0.0 0 1 0.00 E+0 0 0.00 E+0 0 0 1 0.00 E+0 0 0.00 E+0 0 8366 66.67 27 7 2.51 E+0 6 1.85 E+0 5 0 0.0 0 Q1J9 C1.1 27 8 1 1 5.7 6 0.8 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 5.87 E+0 5 0.00 E+0 0 0.00 E+0 0 1956 66.67 #DI V/0 ! Q8IY A2.3 6166 6.667 94 RecName: Full=Intermediate filament protein ifp-1: AltName: Full=Cel IF E1: AltName: Full=Intermediate filament protein E1: Short=IF-E1 RecName: Full=KN motif and ankyrin repeat domaincontaining protein 1: AltName: Full=Ankyrin repeat domain-containing protein 15: AltName: Full=Kidney ankyrin repeatcontaining protein RecName: Full=DNAdirected RNA polymerase subunit beta: Short=RNAP subunit beta: AltName: Full=RNA polymerase subunit beta: AltName: Full=Transcriptase subunit betagi|254765297|sp|B8ZSC 7.1|RPOB_MYCLB RecName: Full=DNAdirected RNA polymerase subunit beta: Short=RNAP subunit beta: AltName: Full=RNA polymerase subunit beta: AltName: Full=Transcriptase subunit beta RecName: Full=Multifunctional CCA protein: Includes: RecName: Full=CCA-adding enzyme: AltName: Full=CCA tRNA nucleotidyltransferase: AltName: Full=tRNA CCApyrophosphorylase: AltName: Full=tRNA adenylyl-/cytidylyltransferase: AltName: Full=tRNA nucleotidyltransferase: AltName: Full=tRNA-NT: Includes: RecName: Full=2'-nucleotidase: Includes: RecName: Full=2',3'-cyclic phosphodiesterase: Includes: RecName: Full=Phosphatase RecName: Full=Putative Fbox/kelch-repeat protein At3g24610 RecName: Full=DNA mismatch repair protein MutSgi|166232143|sp|Q1JJ H0.1|MUTS_STRPC RecName: Full=DNA mismatch repair protein MutS RecName: Full=Coiled-coil domain-containing protein 144C 27 9 1 1 5.7 4 0.5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 28 0 1 1 5.7 4 5.2 28 1 2 1 5.7 3 6.4 28 2 1 1 5.3 4 6.4 28 3 1 1 5.7 1 3 28 4 1 1 5.7 1 28 5 1 1 28 6 2 28 7 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1.71 E+0 6 7.73 E+0 5 1.26 E+0 6 0.00 E+0 0 6.88 E+0 5 4200 00 0.00 E+0 0 1.20 E+0 6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 7.73 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 2576 66.67 2.4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 2.62 E+0 5 5.7 8.2 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 1 5.6 9 1.6 1 1 5.6 8 0.7 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 4.66 E+0 6 0.00 E+0 0 28 8 1 1 5.6 7 1.2 28 9 1 1 5.6 5 2.4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 6.87 E+0 5 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 0 0 4000 00 0 2290 00 95 5700 00 4870 00 #DI V/0 ! #DI V/0 ! #DI V/0 ! P357 24.1 RecName: Full=Manganese resistance protein MNR2 Q5H ZH2. 1 Q9N S85.1 RecName: Full=Ribosome biogenesis protein TSR3 homolog RecName: Full=Carbonic anhydrase-related protein 10: AltName: Full=Carbonic anhydraserelated protein X: Short=CA-RP X: Short=CARP X: AltName: Full=Cerebral protein 15gi|47115606|sp|P61215.1| CAH10_MOUSE RecName: Full=Carbonic anhydrase-related protein 10: AltName: Full=Carbonic anhydraserelated protein X: Short=CA-RP X: Short=CARP Xgi|52000736|sp|Q9N085.1| CAH10_MACFA RecName: Full=Carbonic anhydrase-related protein 10gi|75041009|sp|Q5R4U0. 1|CAH10_PONAB RecName: Full=Carbonic anhydrase-related protein 10 RecName: Full=Carbonic anhydrase-related protein 10 #DI V/0 ! 0.0 0 A0JN 41.1 8733 3.333 #DI V/0 ! P305 01.1 3.16 E+0 5 1053 33.33 #DI V/0 ! O489 15.1 3.63 E+0 6 0.00 E+0 0 0.00 E+0 0 5.63 E+0 6 2763 333.3 #DI V/0 ! #DI V/0 ! Q5IF 00.1 6.27 E+0 6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 2090 000 0 1876 666.7 0 #DI V/0 ! 0.0 0 O431 59.2 Q8R5 08.1 P355 56.3 B3W 6N4.1 RecName: Full=Ribosomal RNA-processing protein 8: AltName: Full=Cerebral protein 1: AltName: Full=Nucleomethylin RecName: Full=HLA class I histocompatibility antigen, Cw-2 alpha chain: AltName: Full=MHC class I antigen Cw*2: Flags: Precursor RecName: Full=Protein NDR1: AltName: Full=Non-race specific disease resistance protein 1: Short=AtNDR1: Flags: Precursor RecName: Full=Autophagyrelated protein 28 RecName: Full=Protocadherin Fat 3: AltName: Full=FAT tumor suppressor homolog 3: Flags: Precursor RecName: Full=Fibrillin-2: Flags: Precursor RecName: Full=Chromosomal replication initiator protein DnaA 29 0 1 1 5.6 4 6.8 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1.15 E+0 6 4.83 E+0 5 0.00 E+0 0 0.00 E+0 0 29 1 1 1 5.6 4 2.4 29 2 1 1 5.6 3 4.4 29 3 1 1 5.6 3 3.9 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 29 4 1 1 5.6 2 1.3 0.00 E+0 0 0.00 E+0 0 29 5 1 1 5.6 1 3 0.00 E+0 0 29 6 1 1 5.6 2 29 7 1 1 5.5 9 5.1 29 8 1 1 5.5 9 2.8 29 9 1 1 5.5 7 2.3 30 0 1 1 5.5 7 30 1 1 1 30 2 1 1 1610 00 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 3.12 E+0 6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q494 20.1 RecName: Full=Probable cysteine desulfurase 1040 000 #DI V/0 ! 0.0 0 Q5R4 P8.1 RecName: Full=Zinc finger protein 574 Q091 79.2 1186 66.67 #DI V/0 ! Q8C XQ6. 1 0.00 E+0 0 2510 000 #DI V/0 ! A1L0 20.1 0.00 E+0 0 0.00 E+0 0 1693 33.33 #DI V/0 ! Q15N V5.1 4.68 E+0 5 0.00 E+0 0 0.00 E+0 0 1560 00 #DI V/0 ! A4W 5D0.1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 O276 98.1 0 0.0 0 Q68D I1.2 RecName: Full=Glutamine synthetase: Short=GS: AltName: Full=Glutamate-ammonia ligase RecName: Full=tRNA (guanine-N(1)-)methyltransferase: AltName: Full=M1Gmethyltransferase: AltName: Full=tRNA [GM37] methyltransferase RecName: Full=RNAbinding protein MEX3A: AltName: Full=RING finger and KH domain-containing protein 4 RecName: Full=3-methyl-2oxobutanoate hydroxymethyltransferase: AltName: Full=Ketopantoate hydroxymethyltransferase: Short=KPHMT RecName: Full=Isocitrate dehydrogenase kinase/phosphatase: Short=IDH kinase/phosphatase: Short=IDHK/P RecName: Full=Anthranilate phosphoribosyltransferase RecName: Full=Zinc finger protein 776 0 3.56 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 7.53 E+0 6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 5.08 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 2.39 E+0 6 2.74 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 5.82 E+0 5 0.00 E+0 0 9133 3.333 0 0.0 0 Q5X G99.2 6.5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 9.69 E+0 5 0.00 E+0 0 0.00 E+0 0 3230 00 #DI V/0 ! Q9B QY9. 3 5.5 6 1.7 0.00 E+0 0 1.15 E+0 6 0.00 E+0 0 3833 33.33 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 A4S6 Y4.1 5.5 6 0.4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 3.22 E+0 6 0.00 E+0 0 0.00 E+0 0 1073 333.3 #DI V/0 ! P981 58.1 0 3833 33.33 1940 00 7966 66.67 96 0 RecName: Full=LysM and putative peptidoglycanbinding domain-containing protein 4 RecName: Full=Dysbindin domain-containing protein 2: Short=Casein kinase-1 binding protein: AltName: Full=CK1BP: AltName: Full=HSMNP1 RecName: Full=Lon protease homolog, mitochondrial: Flags: Precursor RecName: Full=Lowdensity lipoprotein receptorrelated protein 2: Short=LRP-2: AltName: Full=Glycoprotein 330: Short=gp330: AltName: Full=Megalin: Flags: Precursor 30 3 1 1 5.5 3 0.8 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 2.23 E+0 6 0.00 E+0 0 0.00 E+0 0 7433 33.33 #DI V/0 ! Q8IY B8.1 30 4 1 1 5.5 2 0.7 5.5 1 2.7 0.00 E+0 0 2.61 E+0 5 1.10 E+0 6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! #DI V/0 ! O149 24.1 1 0.00 E+0 0 0.00 E+0 0 3666 66.67 1 0.00 E+0 0 0.00 E+0 0 0 30 5 0.00 E+0 0 0.00 E+0 0 30 6 1 1 5.5 1 31.1 0.00 E+0 0 4.70 E+0 6 0.00 E+0 0 1566 666.7 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 P051 09.1 30 7 1 1 5.4 8 1.6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 7.22 E+0 5 0.00 E+0 0 2406 66.67 #DI V/0 ! P487 36.3 30 8 1 1 5.4 8 5.4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 5.10 E+0 6 0.00 E+0 0 0.00 E+0 0 1700 000 #DI V/0 ! P524 24.1 0 97 8700 0 B2IC 30.1 RecName: Full=ATPdependent RNA helicase SUPV3L1, mitochondrial: AltName: Full=Suppressor of var1 3-like protein 1: Short=SUV3-like protein 1: Flags: Precursor RecName: Full=Regulator of G-protein signaling 12: Short=RGS12 RecName: Full=Formate-tetrahydrofolate ligase: AltName: Full=Formyltetrahydrofolate synthetase: Short=FHS: Short=FTHFS RecName: Full=Protein S100-A8: AltName: Full=Calgranulin-A: AltName: Full=Calprotectin L1L subunit: AltName: Full=Cystic fibrosis antigen: Short=CFAG: AltName: Full=Leukocyte L1 complex light chain: AltName: Full=Migration inhibitory factor-related protein 8: Short=MRP-8: Short=p8: AltName: Full=S100 calcium-binding protein A8: AltName: Full=Urinary stone protein band A RecName: Full=Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform: Short=PI3-kinase subunit gamma: Short=PI3K-gamma: Short=PI3Kgamma: Short=PtdIns-3-kinase subunit gamma: AltName: Full=Phosphatidylinositol 4,5-bisphosphate 3-kinase 110 kDa catalytic subunit gamma: Short=PtdIns-3kinase subunit p110gamma: Short=p110gamma: AltName: Full=Phosphoinositide-3kinase catalytic gamma polypeptide: AltName: Full=Serine/threonine protein kinase PIK3CG: AltName: Full=p120-PI3K RecName: Full=Phosphoribosylformyl glycinamidine cyclo-ligase, chloroplastic/mitochondrial: AltName: Full=AIR synthase: Short=AIRS: AltName: Full=Phosphoribosylaminoimidazole synthetase: AltName: Full=VUpur5: Flags: Precursor 30 9 1 1 5.4 8 4.4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 1.58 E+0 5 0.00 E+0 0 0.00 E+0 0 5266 6.667 #DI V/0 ! Q581 75.2 31 0 1 1 5.4 8 0.7 4.73 E+0 6 0.00 E+0 0 0.00 E+0 0 1576 666.7 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q5T HR3. 1 31 1 1 1 5.4 8 6.9 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 3.10 E+0 5 1033 33.33 #DI V/0 ! Q969 X2.1 31 2 1 1 5.4 7 5.2 2.59 E+0 5 0.00 E+0 0 0.00 E+0 0 8633 3.333 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q9N Q79.2 31 3 1 1 5.4 6 2.8 0.00 E+0 0 0.00 E+0 0 2.59 E+0 5 8633 3.333 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 A4W TP4.1 31 4 1 1 5.4 6 4.9 5.4 6 4.2 1 5.4 5 6.1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 6000 00 2 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! 0.0 0 Q38 WL5. 1 Q6M FS1.1 31 6 0.00 E+0 0 1.80 E+0 6 0.00 E+0 0 0.0 0 1 2.94 E+0 5 0.00 E+0 0 0.00 E+0 0 0 1 0.00 E+0 0 0.00 E+0 0 2.30 E+0 5 9800 0 31 5 0.00 E+0 0 0.00 E+0 0 2.35 E+0 5 31 7 1 1 5.4 5 12.4 1 1 5.4 3 1.8 0.00 E+0 0 1.19 E+0 6 0.00 E+0 0 0.00 E+0 0 0 31 8 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 6.22 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 1550 00 3966 66.67 98 0 2073 33.33 0 #DI V/0 ! 0.0 0 D2Y2 83.1 Q571 33.1 B7IF R8.1 RecName: Full=Hydroxylamine reductase: AltName: Full=Hybrid-cluster protein: Short=HCP RecName: Full=EF-hand calcium-binding domaincontaining protein 6: AltName: Full=CAPbinding protein complexinteracting protein 1: AltName: Full=DJ-1binding protein: Short=DJBP RecName: Full=Alpha-Nacetylgalactosaminide alpha-2,6-sialyltransferase 6: AltName: Full=GalNAc alpha-2,6-sialyltransferase VI: AltName: Full=ST6GalNAc VI: Short=ST6GalNAcVI: Short=hST6GalNAc VI: AltName: Full=Sialyltransferase 7F: Short=SIAT7-F RecName: Full=Cartilage acidic protein 1: AltName: Full=68 kDa chondrocyteexpressed protein: Short=CEP-68: AltName: Full=ASPIC: Flags: Precursor RecName: Full=GMP synthase [glutaminehydrolyzing]: AltName: Full=GMP synthetase: AltName: Full=Glutamine amidotransferase RecName: Full=Methionine import ATP-binding protein MetN RecName: Full=Exocyst complex protein EXO70 RecName: Full=Hainantoxin-XVI-13: Short=HNTX-XVI-13: Flags: Precursor RecName: Full=Uncharacterized protein HI_0973 RecName: Full=tRNA(Ile)lysidine synthase: AltName: Full=tRNA(Ile)-2-lysylcytidine synthase: AltName: Full=tRNA(Ile)-lysidine synthetase 31 9 1 1 5.4 3 7.1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 1.51 E+0 6 0.00 E+0 0 0.00 E+0 0 5033 33.33 #DI V/0 ! O190 74.1 32 0 1 1 5.4 2 3.6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 2.68 E+0 5 8933 3.333 #DI V/0 ! P280 28.3 32 1 1 1 5.4 2 0.1 0.00 E+0 0 0.00 E+0 0 2.67 E+0 6 8900 00 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 P785 27.3 32 2 1 1 5.3 9 5.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 2.28 E+0 6 0.00 E+0 0 0.00 E+0 0 7600 00 #DI V/0 ! Q8N GM1. 1 32 3 1 1 5.3 8 2.8 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 2.87 E+0 6 9566 66.67 #DI V/0 ! Q8Y H20.1 32 4 1 1 5.3 7 0.8 1 5.3 7 1.3 0.00 E+0 0 0.00 E+0 0 5.22 E+0 5 6.64 E+0 6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1740 00 1 0.00 E+0 0 0.00 E+0 0 0 32 5 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! #DI V/0 ! B4G BA9. 2 Q6Q7 59.1 32 6 1 1 5.3 2 6.7 1 5.3 2 1.1 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 6.50 E+0 5 0.00 E+0 0 0.00 E+0 0 9.63 E+0 5 0.00 E+0 0 #DI V/0 ! #DI V/0 ! P0DI 62.1 1 0.00 E+0 0 0.00 E+0 0 0 32 7 0.00 E+0 0 0.00 E+0 0 32 8 1 1 5.3 1 3.4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 4.60 E+0 5 0.00 E+0 0 0.00 E+0 0 1533 33.33 #DI V/0 ! Q2N NU1. 1 32 9 1 1 5.3 1 0 1.10 E+0 6 0.00 E+0 0 0.00 E+0 0 3666 66.67 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q91Z U6.1 33 0 1 1 5.3 1 3.5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 5.02 E+0 6 0.00 E+0 0 0.00 E+0 0 1673 333.3 #DI V/0 ! Q927 83.3 0 0 99 2213 333.3 3210 00 2166 66.67 Q9U SI6.1 RecName: Full=Cytidine monophosphate-Nacetylneuraminic acid hydroxylase: Short=CMPN-acetylneuraminic acid hydroxylase: AltName: Full=CMP-Nacetylneuraminate monooxygenase: AltName: Full=CMP-Neu5Ac hydroxylase: AltName: Full=CMP-NeuAc hydroxylase RecName: Full=Serine/threonineprotein kinase B-raf: AltName: Full=Protooncogene B-Raf RecName: Full=DNAdependent protein kinase catalytic subunit: Short=DNA-PK catalytic subunit: Short=DNA-PKcs: AltName: Full=DNPK1: AltName: Full=p460 RecName: Full=Olfactory receptor 4C15: AltName: Full=Olfactory receptor OR11-127: AltName: Full=Olfactory receptor OR11-134 RecName: Full=Beta-(1->2)glucan export ATPbinding/permease protein NdvA RecName: Full=Protein teflon RecName: Full=Spermassociated antigen 17: AltName: Full=Projection protein PF6 homolog RecName: Full=Casparian strip membrane protein 1 RecName: Full=Myosin type-2 heavy chain 1: AltName: Full=Myosin type II heavy chain 1 RecName: Full=E3 ubiquitin-protein ligase IE2: AltName: Full=Immediateearly protein IE2 RecName: Full=Dystonin: AltName: Full=Bullous pemphigoid antigen 1: Short=BPA: AltName: Full=Dystonia musculorum protein: AltName: Full=Hemidesmosomal plaque protein: AltName: Full=Microtubule actin cross-linking factor 2 RecName: Full=Signal transducing adapter molecule 1: Short=STAM-1 33 1 1 1 5.3 7.9 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 0.00 E+0 0 1.14 E+0 5 0.00 E+0 0 3.92 E+0 4 0.00 E+0 0 4.62 E+0 7 0.00 E+0 0 0.00 E+0 0 33 2 1 1 5.2 9 8.9 33 3 1 1 5.2 9 5.7 33 4 2 1 5.2 8 0 4.26 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 1420 00 4.36 E+0 5 0.00 E+0 0 0.00 E+0 0 3.16 E+0 5 0.00 E+0 0 0.00 E+0 0 1453 33.33 1.0 2 A2A SS6.1 33 5 1 1 5.2 8 8.2 1053 33.33 #DI V/0 ! P464 39.3 33 6 2 1 5.2 8 0.5 0.00 E+0 0 1.06 E+0 6 0.00 E+0 0 3533 33.33 0.00 E+0 0 2.65 E+0 6 0.00 E+0 0 8833 33.33 2.5 0 Q86U R5.1 33 7 1 1 5.2 7 4.9 0.00 E+0 0 0.00 E+0 0 5.97 E+0 5 1990 00 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q095 44.1 33 8 1 1 5.2 6 5.6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 5.11 E+0 5 0.00 E+0 0 0.00 E+0 0 1703 33.33 #DI V/0 ! B7L0 43.1 33 9 1 1 5.2 5 13.5 0.00 E+0 0 5.40 E+0 5 0.00 E+0 0 1800 00 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q7V6 D4.1 34 0 1 1 5.2 4 3.8 0.00 E+0 0 0.00 E+0 0 2.79 E+0 5 9300 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q96R 27.2 34 1 1 1 5.2 2 2.8 0.00 E+0 0 0.00 E+0 0 1.10 E+0 6 3666 66.67 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 P136 82.4 0 0 0 100 1540 0000 1306 6.667 3800 0 #DI V/0 ! #DI V/0 ! #DI V/0 ! P709 02.1 A6Q5 99.1 Q9B V10.1 RecName: Full=Variable large protein 15/16: Flags: Precursor RecName: Full=UPF0102 protein NIS_1551 RecName: Full=Dol-PMan:Man(7)GlcNAc(2)-PPDol alpha-1,6mannosyltransferase: AltName: Full=Asparaginelinked glycosylation protein 12 homolog: Short=hALG12: AltName: Full=Dolichyl-PMan:Man(7)GlcNAc(2)-PPdolichyl-alpha-1,6mannosyltransferase: AltName: Full=Mannosyltransferase ALG12 homolog: AltName: Full=Membrane protein SB87 RecName: Full=Titin: AltName: Full=Connectin RecName: Full=Glutathione S-transferase Mu 5: AltName: Full=GST classmu 5: AltName: Full=GSTM5-5 RecName: Full=Regulating synaptic membrane exocytosis protein 1: AltName: Full=Rab-3interacting molecule 1: Short=RIM 1: AltName: Full=Rab-3-interacting protein 2 RecName: Full=ATP synthase subunit delta, mitochondrial: AltName: Full=F-ATPase delta subunit: Flags: Precursor RecName: Full=3-deoxymanno-octulosonate cytidylyltransferase: AltName: Full=CMP-2keto-3-deoxyoctulosonic acid synthase: Short=CKS: Short=CMP-KDO synthase RecName: Full=Aldehyde decarbonylase: Short=AD: AltName: Full=Fatty aldehyde decarbonylase RecName: Full=Olfactory receptor 2M4: AltName: Full=HTPCRX18: AltName: Full=OST710: AltName: Full=Olfactory receptor OR1-55: AltName: Full=Olfactory receptor TPCR100 RecName: Full=Zinc finger protein 35: AltName: Full=Zinc finger protein HF.10 34 2 1 1 5.2 1.2 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 2.37 E+0 6 0.00 E+0 0 7900 00 #DI V/0 ! P511 60.2 34 3 1 1 5.2 4.2 7.4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 Q96S K2.2 5.1 9 0.00 E+0 0 6.33 E+0 5 0.0 0 1 0.00 E+0 0 0.00 E+0 0 0 1 1.41 E+0 6 0.00 E+0 0 4700 00 34 4 0.00 E+0 0 0.00 E+0 0 2110 00 #DI V/0 ! C5A4 B7.1 34 5 1 1 5.1 7 5.8 0.00 E+0 0 0.00 E+0 0 3.76 E+0 5 1253 33.33 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 P467 18.2 34 6 1 1 5.1 7 2 0.00 E+0 0 9.07 E+0 5 0.00 E+0 0 3023 33.33 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q9U S47.2 34 7 1 1 5.1 5 7 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 3.85 E+0 6 0.00 E+0 0 0.00 E+0 0 1283 333.3 #DI V/0 ! Q5B1 31.1 34 8 1 1 5.1 5 6 0.00 E+0 0 0.00 E+0 0 4.72 E+0 5 1573 33.33 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q9B XC0. 1 34 9 1 1 5.1 4 0.3 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 2.39 E+0 6 0.00 E+0 0 7966 66.67 #DI V/0 ! P08F 94.1 35 0 1 1 5.1 4 11.4 2.35 E+0 5 0.00 E+0 0 0.00 E+0 0 7833 3.333 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 Q9N X76.1 35 1 1 1 5.1 3 6.8 5.1 2 2.5 6.49 E+0 6 6.85 E+0 5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! #DI V/0 ! Q581 44.1 1 0.00 E+0 0 0.00 E+0 0 2163 333.3 1 0.00 E+0 0 0.00 E+0 0 0 35 2 0.00 E+0 0 0.00 E+0 0 35 3 1 1 5.1 2 2.8 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 4.95 E+0 5 0.00 E+0 0 0.00 E+0 0 1650 00 0 0 101 2283 33.33 #DI V/0 ! Q0W 486.1 Q6G QV1. 1 RecName: Full=Cone cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha': AltName: Full=cGMP phosphodiesterase 6C: Flags: Precursor RecName: Full=Transmembrane protein 209 RecName: Full=Sadenosylmethionine synthase: Short=AdoMet synthase: AltName: Full=Methionine adenosyltransferase RecName: Full=Programmed cell death protein 2: AltName: Full=Zinc finger protein Rp8 RecName: Full=Putative succinate-semialdehyde dehydrogenase C1002.12c [NADP(+)]: Short=SSDH RecName: Full=Probable carboxypeptidase AN5749: AltName: Full=Peptidase M20 domain-containing protein AN5749: Flags: Precursor RecName: Full=Hydroxycarboxylic acid receptor 1: AltName: Full=G-protein coupled receptor 104: AltName: Full=G-protein coupled receptor 81 RecName: Full=Fibrocystin: AltName: Full=Polycystic kidney and hepatic disease 1 protein: AltName: Full=Polyductin: AltName: Full=Tigmin: Flags: Precursor RecName: Full=CKLF-like MARVEL transmembrane domain-containing protein 6: AltName: Full=Chemokine-like factor superfamily member 6 RecName: Full=Putative rubrerythrin RecName: Full=O-phosphoL-seryl-tRNA:Cys-tRNA synthase 2: AltName: Full=Sep-tRNA:Cys-tRNA synthase 2: Short=SepCysS 2 RecName: Full=Protein FAM196B 35 4 1 1 5.1 2 4.4 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 1.15 E+0 5 0.00 E+0 0 0.00 E+0 0 3833 3.333 #DI V/0 ! Q8T DR2. 2 35 5 1 1 5.0 9 1.3 5.0 7 7.3 0.00 E+0 0 9.34 E+0 4 2.79 E+0 6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 #DI V/0 ! #DI V/0 ! Q140 31.3 1 0.00 E+0 0 0.00 E+0 0 9300 00 1 0.00 E+0 0 0.00 E+0 0 0 35 6 0.00 E+0 0 0.00 E+0 0 35 7 1 1 5.0 6 2.5 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 2.08 E+0 6 0.00 E+0 0 0.00 E+0 0 6933 33.33 #DI V/0 ! Q9N SB2.2 35 8 1 1 5.0 5 9.5 0.00 E+0 0 0.00 E+0 0 2.61 E+0 5 8700 0 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 E9D BV9. 1 35 9 1 1 5.0 5 9.3 0.6 0.00 E+0 0 6.65 E+0 5 0.00 E+0 0 0.00 E+0 0 Q484 45.1 5.0 5 0.00 E+0 0 0.00 E+0 0 0.0 0 1 0.00 E+0 0 0.00 E+0 0 0 1 0.00 E+0 0 0.00 E+0 0 7100 0 36 0 2.13 E+0 5 0.00 E+0 0 2216 66.67 #DI V/0 ! Q8N EZ4.3 36 1 1 1 5.0 4 2.6 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.00 E+0 0 1.26 E+0 6 0.00 E+0 0 4200 00 #DI V/0 ! O147 30.2 36 2 1 1 5.0 2 4.8 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 2.07 E+0 5 0.00 E+0 0 0.00 E+0 0 6900 0 #DI V/0 ! A4G2 T7.1 36 3 1 1 5.0 2 9.6 6.96 E+0 5 0.00 E+0 0 0.00 E+0 0 2320 00 0.00 E+0 0 0.00 E+0 0 0.00 E+0 0 0 0.0 0 C5B HP3. 1 0 0 102 3113 3.333 C6DF 34.1 RecName: Full=Serine/threonineprotein kinase 35: AltName: Full=CLP-36-interacting kinase 1: Short=CLIK-1: AltName: Full=PDLIM1interacting kinase 1: AltName: Full=Serine/threonineprotein kinase 35 L1 RecName: Full=Collagen alpha-6(IV) chain: Flags: Precursor RecName: Full=Ribose-5phosphate isomerase A: AltName: Full=Phosphoriboisomerase A: Short=PRI RecName: Full=Keratin, type II cuticular Hb4: AltName: Full=Keratin-84: Short=K84: AltName: Full=Type II hair keratin Hb4: AltName: Full=TypeII keratin Kb24 RecName: Full=Leucine aminopeptidase 1: AltName: Full=Leucyl aminopeptidase 1: Short=LAP1: Flags: Precursor RecName: Full=UPF0053 protein in cps region: AltName: Full=ORF19 RecName: Full=Histonelysine N-methyltransferase MLL3: AltName: Full=Homologous to ALR protein: AltName: Full=Lysine Nmethyltransferase 2C: Short=KMT2C: AltName: Full=Myeloid/lymphoid or mixed-lineage leukemia protein 3 RecName: Full=Serine/threonineprotein kinase RIO3: AltName: Full=RIO kinase 3: AltName: Full=sudD homolog RecName: Full=tRNA (guanine-N(1)-)methyltransferase: AltName: Full=M1Gmethyltransferase: AltName: Full=tRNA [GM37] methyltransferase RecName: Full=DNA gyrase inhibitor Table S8. Details of the pathways associated with the differentially expressed proteins identified in meningiomas defined by DAVID (A) and PANTHER (B) analysis (A) Summary of DAVID Analysis BIOCARTA Category Term Co unt % pValue BIOCARTA h_intrinsicPathway :Intrinsic Prothrombin Activation Pathway 11 7.74 6479 7.83E14 BIOCARTA h_alternativePathw ay:Alternative Complement Pathway 9 6.33 8028 BIOCARTA h_compPathway:C omplement Pathway 10 7.04 2254 Gen es P02 671, P00 742, P01 042, P00 740, P05 155, P04 070, P01 008, P03 952, P00 748, P00 734, P07 225 7.80E- P01 13 031, P07 357, P00 751, P02 748, P13 671, P01 024, P10 643, P27 918, P06 681 6.24E- P01 12 031, P07 357, P00 751, 103 List Total Pop Hits Po p To tal 14 37 Fold Enrich ment Bonfer roni Benja mini FDR 32 17 29.056 98529 2.19E12 2.19E12 6.55E -11 32 10 14 37 40.415 625 2.18E11 1.09E11 6.52E -10 32 17 14 37 26.415 44118 1.75E10 5.82E11 5.22E -09 BIOCARTA h_classicPathway: Classical Complement Pathway 9 6.33 8028 8.34E12 BIOCARTA h_lectinPathway:L ectin Induced Complement Pathway 8 5.63 3803 7.82E10 BIOCARTA h_amiPathway:Acu te Myocardial Infarction 7 4.92 9577 9.39E08 P02 748, P09 871, P13 671, P01 024, P10 643, P0C 0L5, P06 681 P01 031, P07 357, P02 748, P09 871, P13 671, P01 024, P10 643, P0C 0L5, P06 681 P01 031, P07 357, P02 748, P13 671, P01 024, P10 643, P0C 0L5, P06 681 P02 671, P00 742, P04 070, P01 008, P00 747, P00 734, 104 32 12 14 37 33.679 6875 2.33E10 5.83E11 6.97E -09 32 12 14 37 29.937 5 2.19E08 4.38E09 6.54E -07 32 13 14 37 24.180 28846 2.63E06 4.38E07 7.86E -05 P07 225 BIOCARTA h_extrinsicPathway :Extrinsic Prothrombin Activation Pathway 6 4.22 5352 1.41E06 P02 32 671, P00 742, P04 070, P01 008, P00 734, P07 225 2.81 0.0042 P01 32 6901 12541 031, P13 671, P01 024, P10 643 2.11 0.0147 P00 32 2676 60043 740, P00 747, P00 734 2.11 0.0219 P02 32 2676 52 671, P00 747, P00 734 KEGG_PATHWAY BIOCARTA h_LairPathway:Cel ls and Molecules involved in local acute inflammatory response 4 BIOCARTA h_plateletAppPath way:Platelet Amyloid Precursor Protein Pathway 3 BIOCARTA h_fibrinolysisPath way:Fibrinolysis Pathway 3 Category Term Co unt % pValue Gen es KEGG_PATH WAY hsa04610:Comple ment and coagulation cascades 36 25.3 5211 1.45E55 P01 031, P07 357, P07 358, P04 003, P05 546, P04 070, P08 603, P09 871, P13 105 11 14 37 24.494 31818 3.96E05 5.65E06 0.001 1818 16 14 37 11.226 5625 0.1114 81934 0.0146 66422 3.469 8726 9 14 37 14.968 75 0.3405 58211 0.0452 08605 11.69 7122 11 14 37 12.247 15909 0.4628 63402 0.0602 58352 16.94 674 List Total Pop Hits Fold Enrich ment Bonfer roni Benja mini FDR 57 69 Po p To tal 50 85 46.544 62243 4.64E54 4.64E54 1.26E -52 KEGG_PATH WAY hsa05020:Prion diseases 7 4.92 9577 1.72E06 671, P10 643, P00 751, P01 008, P01 009, P08 697, P01 024, P05 160, P00 734, P01 023, P07 225, P06 681, P02 671, P01 042, P02 748, P07 360, P20 851, P00 742, P05 154, P05 155, P00 740, P05 156, P04 275, P03 952, P00 748, P00 747, P0C 0L5, Q96 IY4 P01 031, P07 357, P07 106 57 35 50 85 17.842 10526 5.51E05 1.84E05 0.001 4912 358, P02 748, P07 360, P13 671, P10 643 0.0155 P02 0365 751, P04 275, P04 004, P07 996 BBID KEGG_PATH WAY hsa04512:ECMreceptor interaction 4 2.81 6901 Category Term Co unt % pValue BBID 79.B_cell_Activati on 2 1.40 8451 0.0737 4001 Category Term Co unt % pValue Gen es PANTHER_P ATHWAY P00011:Blood coagulation 19 13.3 8028 6.96E26 P02 671, P01 042, P05 546, P04 070, P08 519, P00 742, P36 955, P00 740, P01 008, P01 009, P04 275, P08 107 Gen es 57 84 50 85 4.2481 20301 0.8816 01249 0.4134 07129 43.86 9258 List Total Pop Hits Po p To tal 35 8 Fold Enrich ment Bonfer roni Benja mini FDR 19.888 88889 0.4981 24327 0.4981 24327 35.65 0122 Po p To tal 28 57 Fold Enrich ment Bonfer roni Benja mini FDR 34.033 22884 1.04E24 1.04E24 4.84E -23 P01 4 871, P01 857, P01 854 PANTHER_PATHWAY 9 List Total Pop Hits 29 55 697, P03 952, P00 748, P05 160, P01 023, P00 747, P00 734, P07 225 4.22 1.46E- P02 29 5352 06 671, P36 955, P08 697, P00 747, P08 519, Q96 IY4 REACTOME_PATHWAY PANTHER_P ATHWAY P00050:Plasminog en activating cascade 6 Category Term Co unt % PValu e Gen es REACTOME_ PATHWAY REACT_604:Hem ostasis 27 19.0 1408 3.08E17 P04 196, P02 751, P04 114, P04 070, P02 775, P01 008, P09 486, P01 009, P08 697, P05 160, P07 996, P00 734, P01 023, 108 22 28 57 26.868 33856 2.19E05 1.09E05 0.001 015 List Total Pop Hits Fold Enrich ment Bonfer roni Benja mini FDR 54 235 Po p To tal 33 98 7.2297 87234 6.17E16 6.17E16 2.35E -14 REACTOME_ PATHWAY REACT_6900:Sign aling in Immune system 17 11.9 7183 3.54E06 P07 225, P02 671, P02 787, P01 042, P02 768, P00 742, P05 155, P00 740, P10 909, P04 275, P03 952, P00 748, P00 747, P02 647 P02 751, P01 031, P07 357, P07 358, P04 114, P02 748, P04 070, P07 360, P09 871, P13 671, P10 643, P01 834, P00 751, P01 024, P00 734, P06 681, 109 54 286 33 98 3.7403 5224 7.09E05 3.54E05 0.002 6962 P07 225 REACTOME_ PATHWAY REACT_602:Meta bolism of lipids and lipoproteins 8 5.63 3803 0.0079 28057 REACTOME_ PATHWAY REACT_13552:Int egrin cell surface interactions 5 3.52 1127 0.0362 70994 P04 114, P02 649, P02 768, P02 655, P06 727, P02 656, P02 652, P02 647 P02 671, P02 751, P04 275, P04 004, P07 996 54 150 33 98 3.3560 49383 0.1471 68269 0.0516 81004 5.878 1772 54 81 33 98 3.8843 16415 0.5223 62292 0.1686 67706 24.51 1242 (B) Summary of PANTHER Analysis Pathways List Total Count Expected +/- p-value Unclassified 17174 99 109.05 - 1.00E+00 Blood coagulation 57 17 0.36 + 3.79E-21 Plasminogen activating cascade 18 5 0.11 + 2.43E-05 Angiotensin II-stimulated signaling through G proteins and beta-arrestin 38 1 0.24 + 4.30E-02 FAS signaling pathway 38 1 0.24 + 4.50E-02 Integrin signalling pathway 209 1 1.33 - 3.90E-02 p53 pathway 109 1 0.69 + 1.00E+00 Transcription regulation by bZIP transcription factor 58 1 0.37 + 1.00E+00 Nicotinic acetylcholine receptor signaling pathway 105 1 0.67 + 1.00E+00 Muscarinic acetylcholine receptor 2 and 4 signaling pathway 63 1 0.4 + 1.00E+00 Muscarinic acetylcholine receptor 1 and 3 signaling pathway 60 1 0.38 + 1.00E+00 110 Arginine biosynthesis 9 1 0.06 + 1.00E+00 Gonadotropin releasing hormone receptor pathway 282 1 1.79 - 1.00E+00 Inflammation mediated by chemokine and cytokine signaling pathway 289 1 1.84 - 1.00E+00 Huntington disease 164 1 1.04 - 1.00E+00 General transcription regulation 41 1 0.26 + 1.00E+00 Biological Process List Total Count Expected +/- p- value Proteolysis 1131 53 7.18 + 4.04E-30 Complement activation 99 22 0.63 + 4.92E-25 Blood coagulation 271 28 1.72 + 2.43E-23 Response to external stimulus 271 28 1.72 + 2.43E-23 Immune system process 2480 53 15.75 + 2.24E-14 Unclassified 6816 6 43.28 - 1.93E-13 Immune response 725 28 4.6 + 2.46E-12 Response to stimulus 1767 42 11.22 + 3.40E-12 Protein metabolic process 3178 55 20.18 + 3.90E-11 Cell-cell adhesion 724 24 4.6 + 5.69E-09 Cell adhesion 1301 28 8.26 + 1.79E-06 Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process 3782 5 24.02 - 9.89E-05 Metabolic process 8127 77 51.61 + 7.45E-04 Lipid transport 220 9 1.4 + 2.23E-03 Primary metabolic process 7813 73 49.61 + 3.52E-03 Transcription 2224 2 14.12 - 7.65E-03 Transcription from RNA polymerase II promoter 2214 2 14.06 - 8.14E-03 Molecular Function List Total Count Expected +/- p- value Lipid transporter activity 102 24 0.65 + 4.23E-28 Serine-type peptidase activity 350 30 2.22 + 6.57E-23 Serine-type endopeptidase inhibitor activity 125 21 0.79 + 2.10E-21 Peptidase inhibitor activity 196 22 1.24 + 8.82E-19 Peptidase activity 717 33 4.55 + 3.40E-17 Enzyme inhibitor activity 389 22 2.47 + 1.17E-12 Metallopeptidase activity 233 17 1.48 + 3.16E-11 Hydrolase activity 2223 40 14.12 + 8.57E-08 Unclassified 7724 18 49.05 - 1.87E-07 Transporter activity 1049 25 6.66 + 1.55E-06 Protein binding 3073 44 19.51 + 9.98E-06 Nucleic acid binding 3715 4 23.59 - 2.24E-05 Enzyme regulator activity 1216 23 7.72 + 3.66E-04 Calmodulin binding 279 10 1.77 + 1.97E-03 DNA binding 2309 2 14.66 - 3.93E-03 Calcium-dependent phospholipid binding 139 7 0.88 + 5.04E-03 Transcription factor activity 2041 2 12.96 - 2.05E-02 111 Transcription regulator activity 2041 2 12.96 - 2.05E-02 Transferase activity 1601 1 10.17 - 4.49E-02 Cellular Component List Total Count Expected +/- p- value Extracellular region 601 18 3.82 + 2.04E-06 Intermediate filament cytoskeleton 87 5 0.55 + 9.18E-03 Extracellular matrix 558 11 3.54 + 3.28E-02 Unclassified 17814 101 113.12 - 4.34E-02 PANTHER Protein Class List Total Count Expected +/- p- value Apolipoprotein 102 24 0.65 + 5.10E-28 Serine protease 350 30 2.22 + 7.94E-23 Complement component 90 20 0.57 + 1.33E-22 Transfer/carrier protein 475 32 3.02 + 2.30E-21 Serine protease inhibitor 125 21 0.79 + 2.54E-21 Protease inhibitor 196 22 1.24 + 1.07E-18 Protease 717 33 4.55 + 4.11E-17 Defense/immunity protein 694 30 4.41 + 1.26E-14 Metalloprotease 233 17 1.48 + 3.82E-11 Hydrolase 1852 40 11.76 + 3.95E-10 Unclassified 6898 11 43.8 - 1.96E-09 Cell adhesion molecule 668 19 4.24 + 9.40E-06 Nucleic acid binding 2724 3 17.3 - 2.31E-03 Intracellular calcium-sensing protein 279 10 1.77 + 2.38E-03 Calmodulin 279 10 1.77 + 2.38E-03 Annexin 139 7 0.88 + 6.09E-03 Enzyme modulator 1540 23 9.78 + 1.85E-02 Transcription factor 2041 2 12.96 - 2.47E-02 Intermediate filament 87 5 0.55 + 4.59E-02 112 Table S9. ELISA-based measurement of serum HPX, Apo E, Apo A1 and RBP in healthy controls and different grades of meningioma patients A. ELISA-based measurement of serum HPX HC HC 01 HC 02 HC 03 HC 04 HC 05 HC 06 HC 07 HC 08 HC 09 HC 10 HC 11 HC 12 HC 13 HC 14 HC 15 HC 16 HC 17 HC 18 HC 19 HC 20 HC 21 HC 22 HC 23 Conc. (g/L) 1.21 1.09 0.89 0.78 0.83 0.68 1.23 1.07 1.06 1.31 0.82 0.67 1.33 1.32 0.98 0.49 1.06 1.28 0.84 0.93 1.18 1.21 0.97 HC HC 24 HC 25 HC 26 HC 27 HC 28 HC 29 HC 30 HC 31 HC 32 HC 33 HC 34 HC 35 HC 36 HC 37 HC 38 HC 39 HC 40 HC 41 HC 42 HC 43 HC 44 HC 45 Conc. (g/L) 1.18 1.07 1.02 0.97 0.69 0.85 0.7 0.49 0.93 1.11 1.23 1.05 0.89 1.07 1.02 0.68 0.7 0.67 0.91 1.23 1.17 1.08 MG I MG I (1) MG I (2) MG I (3) MG I (4) MG I (5) MG I (6) MG I (7) MG I (8) MG I (9) MG I (10) MG I (11) MG I (12) MG I (13) MG I (14) Conc. (g/L) 1.71 1.03 1.16 0.59 2.19 0.56 0.91 1.8 1.43 1.37 0.67 1.78 1.87 1.61 MG II Conc. (g/L) MG II (1) MG II (2) MG II (3) MG II (4) MG II (5) MG II (1) 1.04 2.63 2.68 2.43 3.71 1.04 MG III* MG III (1) MG III (2) MG III (3) Conc. (g/L) 3.7 3.65 3.82 HC MG I MG II MG III* Number of subjects 45 14 5 3 Minimum 0.49 0.56 1.04 3.65 25% Percentile 0.83 0.85 1.74 3.65 Median 1.02 1.4 2.63 3.7 75% Percentile 1.18 1.78 3.19 3.82 113 Maximum 1.33 2.19 3.71 3.82 Mean 0.97 1.33 2.49 3.72 Std. Deviation 0.22 0.52 0.95 0.09 Std. Error of Mean 0.03 0.14 0.43 0.05 B. ELISA-based measurement of serum Apo E HC Conc. (mg/L) HC Conc. (mg/L) MG I Conc. (mg/L) MG II Conc. (mg/L) HC 01 HC 02 HC 03 HC 04 HC 05 HC 06 HC 07 HC 08 HC 09 HC 10 HC 11 HC 12 HC 13 HC 14 HC 15 HC 16 HC 17 HC 18 HC 19 HC 20 HC 21 HC 22 HC 23 42.78 56.09 146.1 65.09 151.24 49.87 157.67 102.6 43.89 149.08 60.45 147.09 42.89 40.87 60.46 77.89 89.76 49.8 56.03 48.1 53.95 48.03 65.23 HC 24 HC 25 HC 26 HC 27 HC 28 HC 29 HC 30 HC 31 HC 32 HC 33 HC 34 HC 35 HC 36 HC 37 HC 38 HC 39 HC 40 HC 41 HC 42 HC 43 HC 44 HC 45 49.48 50.9 59.4 50.62 60.62 44.89 47.98 102.9 101.78 98.09 45.02 126.9 62.8 50.75 49.3 52.87 70.64 80.67 180.91 121.23 51.17 101.08 MG I (1) MG I (2) MG I (3) MG I (4) MG I (5) MG I (6) MG I (7) MG I (8) MG I (9) MG I (10) MG I (11) MG I (12) MG I (13) MG I (14) 120.56 109.45 98.58 167.89 122.74 112.03 72.61 83.26 184.35 138.76 148.03 157.3 43.29 155.04 MG II (1) MG II (2) MG II (3) MG II (4) MG II (5) MG II (1) 106.23 176.05 61.87 92.99 183.77 106.23 MG III* MG III (1) MG III (2) MG III (3) Conc. (mg/L) 109.67 112.78 119.67 HC MG I MG II MG III* Number of subjects 45 14 5 3 Minimum 40.87 43.29 61.87 109.7 25% Percentile 49.64 94.75 77.43 109.7 114 Median 60.45 121.7 106.2 112.8 75% Percentile 101.4 155.6 179.9 119.7 Maximum 180.9 184.4 183.8 119.7 Mean 77 122.4 124.2 114 Std. Deviation 38.13 39.45 53.43 5.118 Std. Error of Mean 5.684 10.54 23.89 2.955 C. ELISA-based measurement of serum Apo AI HC Conc. (g/L) HC Conc. (g/L) MG I Conc. (g/L) MG II Conc. (g/L) HC 01 HC 02 HC 03 HC 04 HC 05 HC 06 HC 07 HC 08 HC 09 HC 10 HC 11 HC 12 HC 13 HC 14 HC 15 HC 16 HC 17 HC 18 HC 19 HC 20 HC 21 HC 22 HC 23 1.5 1.32 1.11 0.98 1.54 1.26 0.94 1.37 1.27 1.18 0.78 1.17 1.16 1.03 1.42 1.35 0.86 0.52 1.32 1.18 1.67 1.03 0.79 HC 24 HC 25 HC 26 HC 27 HC 28 HC 29 HC 30 HC 31 HC 32 HC 33 HC 34 HC 35 HC 36 HC 37 HC 38 HC 39 HC 40 HC 41 HC 42 HC 43 HC 44 HC 45 0.56 1.26 1.45 1.34 1.29 1.41 1.17 1.21 1.39 1.22 1.18 1.27 1.15 0.73 0.94 1.03 1.12 0.67 0.91 1.23 1.17 1.08 MG I (1) MG I (2) MG I (3) MG I (4) MG I (5) MG I (6) MG I (7) MG I (8) MG I (9) MG I (10) MG I (11) MG I (12) MG I (13) MG I (14) 3.19 2.53 3.82 0.92 0.95 1.52 0.81 0.58 2.12 2.31 0.48 0.75 2.21 2.91 MG II (1) MG II (2) MG II (3) MG II (4) MG II (5) 8.04 2.63 7.68 4.43 6.71 MG III* MG III (1) MG III (2) MG III (3) Conc. (g/L) 115 15.67 14.98 15.78 HC MG I MG II MG III* Number of subjects 45 14 5 3 Minimum 0.52 0.48 2.63 14.98 25% Percentile 1.005 0.795 3.53 14.98 Median 1.18 1.82 6.71 15.67 75% Percentile 1.32 2.63 7.86 15.78 Maximum 1.67 3.82 8.04 15.78 Mean 1.15 1.79 5.89 15.48 Std. Deviation 0.25 1.08 2.30 0.43 Std. Error of Mean 0.038 0.29 1.03 0.25 D. ELISA-based measurement of serum plasma RBP HC Conc. (mg/L) HC Conc. (mg/L) MG I Conc. (mg/L) MG II Conc. (mg/L) HC 01 HC 02 HC 03 HC 04 HC 05 HC 06 HC 07 HC 08 HC 09 HC 10 HC 11 HC 12 HC 13 HC 14 HC 15 HC 16 24.05 29.8 45.98 52.56 33.98 21.9 29.89 37.02 67.89 55.43 27.56 23.98 33.05 31.78 33.02 28.97 12.78 13.09 41.94 18.56 32.93 HC 24 HC 25 HC 26 HC 27 HC 28 HC 29 HC 30 HC 31 HC 32 HC 33 HC 34 HC 35 HC 36 HC 37 HC 38 HC 39 39.78 50.02 47.43 36.56 31.9 29.78 32.78 45.09 48.67 56.9 65.09 45.9 43.01 36.89 38.32 49.45 52.31 80.67 80.91 21.23 51.17 MG I (1) MG I (2) MG I (3) MG I (4) MG I (5) MG I (6) MG I (7) MG I (8) MG I (9) MG I (10) MG I (11) MG I (12) MG I (13) MG I (14) 36.56 31.9 29.78 32.78 45.09 48.67 56.9 65.09 45.9 43.01 36.89 38.32 49.45 52.31 MG II (1) MG II (2) MG II (3) MG II (4) MG II (5) MG II (1) 56.23 36.05 61.87 62.99 53.77 56.23 MG III* MG III (1) MG III (2) MG III (3) Conc. (mg/L) HC 17 HC 18 HC 19 HC 20 HC 21 HC 40 HC 41 HC 42 HC 43 HC 44 116 43.56 35.89 41.67 HC 22 HC 23 36.73 41.78 HC 45 31.08 HC MG I MG II MG III* Number of subjects 45 14 5 3 Minimum 12.78 29.78 36.05 35.89 25% Percentile 29.85 35.62 44.91 35.89 Median 36.89 44.05 56.23 41.67 75% Percentile 49.06 50.17 62.43 43.56 Maximum 80.91 65.09 62.99 43.56 Mean 39.77 43.76 54.18 40.37 Std. Deviation 15.31 10.19 10.84 4.00 Std. Error of Mean 2.28 2.72 4.85 2.31 * MG III sample was analyzed in three technical replicates 117 Table S10. Statistical summary of ROC curve analysis for evaluating performance of different serum proteins for prediction of grade I and grade II meningiomas Classifier protein HC vs. MG I Apolipoprotein E (Apo E) Hemopexin (HPX) Apolipoprotein A-I (Apo A-I) Plasma retinolbinding protein (RBP4) HC vs. MG II MG I vs. MG II AUC 95% CI AUC 95% CI AUC 95% CI 0.794 0.654 - 0.933 0.813 0.647-0.979 0.514 0.164-0.864 0.699 0.616 0.492 - 0.906 0.383 - 0.849 0.906 0.937 0.738 - 1.07 0.783 - 0.987 0.871 0.957 0.636-0.987 0.862-1.052 0.608 0.457 - 0.759 0.804 0.624 - 0.985 0.757 0.479- 0.94 Table S11. Comparison of the fold changes of the differentially expressed proteins identified from iTRAQ data with proteins reported in published literature in tissue and CSF samples Name of Protein Vitamin D-binding protein Apolipoprotein A-I α-1-antitrypsin iTRAQ results obtained from Q-TOF data Benign Atypical Anaplastic 0.85 1.2 0.78 2.28 1.69 Name of Protein Serum albumin precursor Apolipoprotein E α-1-antitrypsin Transthyretin 7.09 6.24 Meningioma tissue proteomics data17 13.26 7.33 Benign 0.735 Atypical 0.374 Anaplastic 0.628 3.951 1.841 1.87 1.067 1.937 2.349 iTRAQ results obtained from QTOF data Meningioma CSF proteomics data20 Benign Atypical Anaplastic Meningioma vs. Non brain tumor samples 0.63 2.39 1 1 1.05 1.48 6.24 1.42 0.61 2.03 7.33 1.86 4.51 2.87 2.73 -3.1 118