BioSci 145A Lecture 13 - 2/20/2001
Transcription factors II
•
Topics we will cover today
– implications of transgenic technology
– Principles of gene regulation
– Identification of regulatory elements
– Identification of regulatory element binding proteins
– Functional analysis
– Transcription factors - introduction
– Modulation of transcription factor activity
– Tour of molecular interaction screening facility
•
transcription factor resources
– http://transfac.gbf-braunschweig.de/TRANSFAC/
– http://bioinformatics.weizmann.ac.il/transfac/
• detailed transcription factor database
– http://copan.bioz.unibas.ch/homeo.html
• collected information about homeobox genes
– http://biochem1.basic-ci.georgetown.edu/nrr/nrr.html
• nuclear receptor resource
•
references
– nuclear transport Nakielny and Dreyfuss (1999) Cell
99, 677-690.
– Nuclear pore structure Daneholt (1997) Cell 88, 585588.
BioSci 145A lecture 13
1
©copyright
Bruce Blumberg 2000. All rights reserved
Gene transfer technology - implications
•
•
Genetics and reverse genetics
– gene transfer and selection technology speeds up
genetic analysis by orders of magnitude
– virtually all conceivable experiments are now
possible
• all questions are askable
– much more straightforward to understand gene
function using knockouts and transgenics
• gene sequences are coming at an unprecedented
rate from the genome projects
• Knockouts and transgenics remain very
expensive to practice
– other yet undiscovered technologies will be
required to understand gene function.
Clinical genetics
– Molecular diagnostics are becoming very widespread
as genes are matched with diseases
• huge growth area for the future
• big pharma is dumping billions into diagnostics
– room for great benefit and widespread abuse
• diagnostics will enable early identification and
treatment of diseases
• but insurance companies will want access to
these data to maximize profits
BioSci 145A lecture 13
2
©copyright
Bruce Blumberg 2000. All rights reserved
Gene transfer technology - implications (contd)
•
•
•
gene therapy
– new viral vector technology is making this a reality
• now possible to get efficient transfer and
reasonable regulation
– long lag time from laboratory to clinic, still working
with old technology in many cases
protein engineering
– not as widely appreciated as more glamorous
techniques such as gene therapy and transgenic crops
– better drugs, eg more stable insulin, TPA for heart
attacks and strokes, etc.
– more efficient enzymes (e.g. subtilisin in detergents)
– safe and effective vaccines
• just produce antigenic proteins rather than using
inactivated or attenuated organisms to reduce
undesirable side effects
metabolite engineering
– enhanced microbial synthesis of valuable products
• eg indigo (jeans)
• vitamin C
– generation of entirely new small molecules
• transfer of antibiotic producing genes to related
species yields new antibiotics (badly needed)
– reduction of undesirable side reactions
• faster more efficient production of beer
BioSci 145A lecture 13
3
©copyright
Bruce Blumberg 2000. All rights reserved
Gene transfer technology - implications (contd)
•
•
transgenic food
– gene transfer techniques have allowed the creation of
desirable mutations into animals and crops of
commercial value
• disease resistance (various viruses)
• pest resistance (Bt cotton)
• pesticide resistance
• herbicide and fungicide resistance
• growth hormone and milk production
– effective but necessary?
– negative implications
• pesticide and herbicide resistance lead to much
higher use of toxic compounds
• results are not predictable due to small datasets
• at least one herbicide (bromoxynil) for which
resistance was engineered has since been banned
plants as producers of specialty chemicals
– still very underutilized since plant technology yet
lags behind techniques in animals
– great interest in using plants as factories to produce
materials more cheaply and efficiently
• especially replacements for petrochemicals
– plants and herbs are the original source of many
pharmaceutical products hence it remains possible to
engineer them to overproduce desirable substances
BioSci 145A lecture 13
4
©copyright
Bruce Blumberg 2000. All rights reserved
Principles of gene regulation
•
•
Why does gene expression need to be controlled anyway?
– Primary purpose in multicellular organisms is to
execute precise developmental decisions so that
• proper genes are expressed at
– appropriate time
– correct place
– at the required levels
• so that development, growth and differentiation
proceed correctly
– maintenance of homeostasis
• produce required substances in appropriate amounts
– nutrients, cofactors, etc.
• degrade undesired substances from
– diet
– metabolism
– injury
• inter and intracellular signaling processes
Where are the control points?
– Activation of gene structure
– initiation of transcription
– processing of the transcript to mRNA
– transport of mRNA to cytoplasm
– translation of mRNA
– processing and stability of protein
BioSci 145A lecture 13
5
©copyright
Bruce Blumberg 2000. All rights reserved
Principles of gene regulation (contd)
•
Activation of gene structure
– genes are active only in cells where they are
expressed
– structure of gene determines whether it is can be
transcribed or not
– activation of an active structure may be one of the
first steps in gene regulation
• modification of DNA
– methylation of DNA inactivates genes
– active genes are hypomethylated
• modification of histones
– methylation and acetylation of histones
activates gene expression
» acetylase activates
• active genes are in an open, hypomethylated
coformation. associated histones are
hyperracetylated
– one of the primary responsibilities of cell-type
specific transcription factors is to facilitate the
formation of an active chromatin conformation
• majority of alleged co-activator and co-repressor
proteins are relatively non-specific modifiers of
chromatin conformation that interact with
specific factors targeting chromatin remodeling
BioSci 145A lecture 13
6
©copyright
Bruce Blumberg 2000. All rights reserved
Principles of gene regulation (contd)
BioSci 145A lecture 13
7
©copyright
Bruce Blumberg 2000. All rights reserved
Principles of gene regulation (contd)
•
Initiation of transcription
– Once the DNA template is accessible, the next
requirement is to form the initiation complex
• although other forms of regulation are important,
the majority of regulatory events occur at the
initiation of transcription
– genes under common control share response
elements (aka cis-cting elements, enhancers)
• these sequences are presumed to be recognized
by specific protein(s)
• the protein(s) functions as a transcription factor
needed for RNA polymerase to initiate
• the active protein is only available when the
gene is to be expressed
– response elements are often cell-type or tissuespecific
• because binding proteins are cell-type specific
• but this is a tautology
– each gene has multiple response elements
• each regulatory event depends on the binding of
a protein to a particular response element
• any one of these can independently activate the
gene
• combinatorial regulation by multiple elements
and proteins is a central mechanism by which
levels of gene expression are modulated
BioSci 145A lecture 13
8
©copyright
Bruce Blumberg 2000. All rights reserved
Principles of gene regulation (contd)
– cis-acting control elements can be located many
kilobases away from the transcriptional start site
• in intergenic regions
• in introns
• some elements may be quite close to TATA box
or other intitiator elements
– cis-acting elements are responsible for allowing the
recruitment of TBP and assembly of the initiation
complex.
BioSci 145A lecture 13
9
©copyright
Bruce Blumberg 2000. All rights reserved
Transcription factors and the preinitiation complex
•
Model for cooperative assembly of an activated transcription initiation
complex at the TTR promoter in hepatocytes.
– Four activators enriched in hepatocytes plus ubiquitous AP-1 factors
bind to sites in the hepatocyte-specific enhancer and promoterproximal region of the TTR gene
– Activation domains of the bound activators interact extensively with
co-activators, TAF subunits of TFIID,Srb/mediator proteins and
general transcription factors. This causes looping of DNA and
formation of stable initiation complex
– Highly cooperative nature of complex assembly prevents initiation
complex from forming in other cells that lack all four of the
hepatocyte-enriched transcription factors.
BioSci 145A lecture 13
10
©copyright
Bruce Blumberg 2000. All rights reserved
Principles of gene regulation (contd)
•
processing of the transcript to mRNA
– RNA is synthesized as an exact copy of DNA
• heterogeneous nuclear RNA (hnRNA)
– hnRNA gets capped and polyadenylated
– introns are spliced out by the spliceosome, a large
complex of RNA and proteins.
• exons can also be spliced out as well. Alternative
splicing may produce proteins with new functions.
– Molecular mechanisms underlying alternative
splicing are still only poorly understood
– regulation of alternative splicing is important in
the CNS and for sex determination
– splice junctions are read in pairs
• spliceosome binds to a 5’ splice donor and scans for
a lariat sequence followed by a 3’ splice acceptor
• mutations in either site can lead to exon skipping
– principle underlying gene trapping
– mRNA is now ready for transport to cytoplasm
– some organisms perform trans splicing between mRNAs
• another way to generate mRNA diversity
BioSci 145A lecture 13
11
©copyright
Bruce Blumberg 2000. All rights reserved
Principles of gene regulation (contd)
•
transport of mRNA to cytoplasm
– capping, polyadenylation and
splicing of mRNA are
prerequisites to transport
– macromolecules are specifically
transported bidirectionally though
nuclear pores
• direction controlled by
nuclear import and export
signals in macromolecules
– fully processed mRNAs are
packaged into ribonucleoprotein
particles, mRNPs
• hnRNP proteins contain
nuclear export sequences
– These are transported through the
pore complex, unwinding as they
do so
– On the cytoplasmic side of the
pore, the mRNA is stripped from
the RNP by binding to ribosomes
– those with signal sequences are
paused and subsequently
associate with ER
– those without are translated
directly
BioSci 145A lecture 13
12
©copyright
Bruce Blumberg 2000. All rights reserved
Principles of gene regulation (contd)
•
translation of mRNA
– by default, mRNAs are all translated
– efficiency of translation is important for protein
levels.
• regulatory genes tend to be poorly translated
– two primary mediators of efficiency
• consensus around the ATG
– optimum is ACCACCATGG
– most important factor is a G following
ATG (A gives about 40% of protein
– underlined sequence will give very high
levels of translation - NcoI site
• stability of mRNA in the cytoplasm varies
– many short lived mRNAs have multiple
copies of the sequence AUUUA in 3’ UTR
BioSci 145A lecture 13
13
©copyright
Bruce Blumberg 2000. All rights reserved
Principles of gene regulation (contd)
• stability of mRNA (contd)
– others mRNAs are specifically degraded,
e.g. transferrin
– in the absence of iron, a specific protein
(IRE-BP) binds to a region of the
transferring mRNA containing AUUUA
sequences
– this protects the mRNA from degradation,
transferrin is synthesized and iron
accumulates
– iron binds to IRE-BP and dissociates it
from mRNA
» AUUUA mediates degradation
BioSci 145A lecture 13
14
©copyright
Bruce Blumberg 2000. All rights reserved
Identification of regulatory elements
•
Given a gene of interest, how does one go about studying
its regulation?
– First step is to isolate cDNA and genomic clones.
– Map cDNA to genomic sequence
• identify introns, exons
• locate approximate transcriptional start
– recognizing elements, e.g. TATA box
– 5’ primer extension or nuclease mapping
• get as much 5’ and 3’ flanking sequence as is
possible
– fuse largest chunk of putative promoter you can get
to a suitable reporter gene.
– Test whether this sequence is necessary and
sufficient for correct regulation
• how much sequence is required for correct
regulation?
– what is correct regulation?
» In cultured cells
» in animals?
– typical result is the more you look, the more
you find.
• questions are usually asked specifically. That is,
what part of the putative promoter is required for
activity in cultured liver cells?
– doesn’t always hold in vivo.
BioSci 145A lecture 13
15
©copyright
Bruce Blumberg 2000. All rights reserved
Identification of regulatory elements (contd)
•
Promoter mapping
– nuclease footprinting of promoter to identify regions
that bind proteins
– make various deletion constructs
• Previously made by ExoIII deletions or insertion
of linkers (linker scanning)
• typical method today is to PCR parts of the
promoter and clone into a promoterless reporter
– map activity of promoter related to deletions
• incremental changes in activity indicate regions
important for activity
– test elements for activity
BioSci 145A lecture 13
16
©copyright
Bruce Blumberg 2000. All rights reserved
Identification of binding proteins
•
•
How to identify what factors bind to putative elements?
– examine the sequence
• does it contain known binding sites?
• if yes, do such proteins bind to the isolated
element in gel-shift experiments?
– do the elements bind proteins from nuclear extracts?
• gel shift (EMSA) experiments
– clone the elements into reporters with minimal
promoters.
• do these constructs recapitulate activity?
Biochemical purification of binding proteins
– tedious, considerable biochemical skill required
– two basic approaches
• fractionate nuclear extracts chromatographically
and test fractions for ability to bind the element in
EMSA
• DNA-affinity chromatography
– multimerize the element and bind to a resin
– pass nuclear extracts across column and
purify specific binding proteins
– protein microsequencing
– predict DNA sequence from amino acid sequence
• look in the database
• prepare oligonucleotides and screen library
BioSci 145A lecture 13
17
©copyright
Bruce Blumberg 2000. All rights reserved
Identification of binding proteins (contd)
•
•
Biochemical purification of binding proteins (contd)
– advantages
• gold standard
• if you can purify proteins, this will always work
– disadvantages
• slow, tedious
• need good protein sequencing facility
• biochemical expertise required
• expense of preparing preparative quantities of
nuclear extracts
Molecular biological approaches
– oligonucleotide screening of expression libraries
(Singh screening)
• multimerize oligonucleotide and label with 32P
• screen expression library to identify binding
proteins
• advantages
– straightforward
– much less biochemical expertise required
– relatively fast
• disadvantages
– can’t detect binding if multiple partners are
required
– fair amount of “touch” required
BioSci 145A lecture 13
18
©copyright
Bruce Blumberg 2000. All rights reserved
Identification of binding proteins (contd)
•
Molecular biological approaches (contd)
– yeast one-hybrid assay
• clone element of interest into a reporter construct
(e.g. -gal) and make stable yeast strain
• transfect in aliquots of cDNA expression
libraries that have fragments of DNA fused to
yeast activator
• if the fusion protein binds to your element then
the reporter gene will be activated
• advantages
– somewhat more of a functional approach
– eukaryotic milieu allows some protein
modification
• disadvantages
– slow, tedious purification of positives
– can’t detect dimeric proteins
– sensitivity is not so great
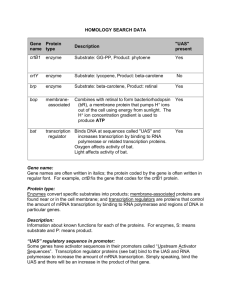
AD
His
Bait elements
BioSci 145A lecture 13
19
©copyright
lacZ
Reporter(s)
Bruce Blumberg 2000. All rights reserved
Identification of binding proteins (contd)
•
Molecular biological approaches (contd)
– expression cloning (sib screening)
• clone element of interest (or promoter) into a
suitable reporter construct (e.g. luciferase)
• transfect (or inject, or infect, etc) pools (~10,000
cDNAs each) of cDNA expression libraries and
assay for reporter gene
• retest positive pools in smaller aliquots (~1000)
• repeat until a pure cDNA is found
– advantages
– functional approach
– presumably using the appropriate cell type
so modifications occur
– possibility to detect dimers with
endogenous proteins
– disadvantages
• VERY TEDIOUS
• very slow, much duplication in pools, extensive
rescreening is required
• could be expensive
BioSci 145A lecture 13
20
©copyright
Bruce Blumberg 2000. All rights reserved
Identification of binding proteins (contd)
– in vitro expression cloning (IVEC)
• transcribe and translate cDNA libraries in vitro
into small pools of proteins (~100)
• EMSA to test protein pools for element binding
• unpool cDNAs and retest
• advantages
– functional approach
– smaller pools increase sensitivity
• disadvantages
– can’t detect dimers
– very expensive (TNT lysate)
– considerable rescreening still required
– tedious, countless DNA minipreps required
BioSci 145A lecture 13
21
©copyright
Bruce Blumberg 2000. All rights reserved
Identification of binding proteins (contd)
– hybrid screening system 1
• begin with cDNA libraries in 384-well plates, 1
cDNA per well
• pool cDNAs using robotic workstation
• prepare DNA with robotic workstation
• transcribe and translate protein in vitro
• test for ability to bind DNA element using
sensitive, high-throughput assay
– fluorescence
– radioactive assay
• retest components of positive pools
• advantages
– very fast, only two steps required, ~ 2
weeks
– little work required
• disadvantages
– expense of robotics
– won’t detect dimers (unless 1 partner
known)
– expense of reagents (TNT, radionuclides,
fluorescent labels
BioSci 145A lecture 13
22
©copyright
Bruce Blumberg 2000. All rights reserved
Identification of binding proteins (contd)
•
– hybrid screening system 2
• prepare reporter cell line with element or
promoter driving reporter gene (e.g. luciferase)
• prepare cDNA pools as in system 1
• use robotic workstation to transfect cDNA
libraries into reporter cells
• assay for reporter gene
• advantages
– very fast
– truly functional approach
– use of cells allows modifications
– can detect dimers if one partner is already
present in cell
• disadvantages
– expense of equipment
OK, you have your element and binding protein, now
what?
– functional analysis depends on type of protein you
are dealing with
– goal will be to prove that this protein is necessary
and sufficient to confer regulation onto the promoter,
in vivo
• many just stop at works on the element
BioSci 145A lecture 13
23
©copyright
Bruce Blumberg 2000. All rights reserved
Transcription factors bind to regulatory elements
•
•
The response element binding proteins you have carefully
identified are transcription factors.
– There are many types. The primary mode of
classification is via the type of DNA-binding
domains and intermolecular interactions (next time)
Features of transcription factors
– typically these proteins have multiple functional
domains
• can frequently be rearranged or transferred
– DNA-binding domains
• these domains take many forms that will be
discussed next time
• see also the list in TRANSFAC
http://transfac.gbf-braunschweig.de/TRANSFAC/
– Activation domains
• these are polypeptide sequences that activate
transcription when fused to a DNA-binding
domain
• these are diverse in sequence, 1% of random
sequences fused to GAL4 can activate
• many activation domains are rich in acidic
residues and assume an amphipathic -helix
conformation when associated with coactivator
proteins
• interact with histone acetylases that destabilize
nucleosomes and open chromatin
BioSci 145A lecture 13
24
©copyright
Bruce Blumberg 2000. All rights reserved
Transcription factors bind to regulatory elements (contd)
•
Features of transcription factors (contd)
– repression domains
• functional converse of activation domains
• short and diverse in amino acid sequence
– some are rich in hydrophobic aa
– others are rich in basic aa
• some interact with proteins having histone
deacetylase activity, stabilizes nucleosomes and
condenses chromatin
• others compete with activators for the same
sequence and contacts with the transcription
machinery
– protein:protein interaction domains
• these are diverse in sequence but do contain
structural motifs
• leucine zipper
• helix-loop-helix
BioSci 145A lecture 13
25
©copyright
Bruce Blumberg 2000. All rights reserved