Honours Genetics Research Tutorial
advertisement

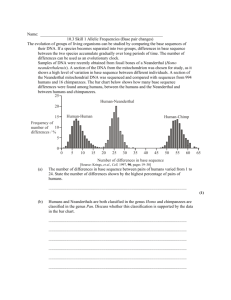
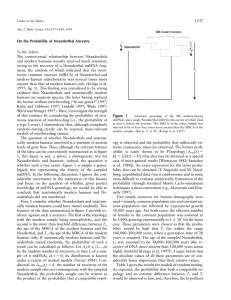
Molecular Anthropology Genetics Research Tutorial 1 www.abdn.ac.uk/~gen155/lectures/restut.ppt Human Origins Mya 5 0.1 4 3 Australopithecus 2 1 Early Homo sp 0 H sapiens Neanderthals Mya = million years ago Birth of Jesus was 0.002 Mya European Human Prehistory • Most of European history is prehistory • We (modern-type humans) got here about 40000 years ago: the Palaeolithic (old stone age) - primarily hunter-gatherer economy displacing the indigenous Neanderthal type humans • Farming spread across Europe from the Middle East (“Fertile Crescent”) from around 7000 Ya - Neolithic (new stone age) European time chart Neanderthal, Cro-Magnon and modern Low forehead, prominent eyebrow ridges Very similar to us Paleolithic Cave Art from Lascaux (Dordogne, France) (About 17000 Ya) The mitochondrial chromosome • About 15kb of mtDNA • Mutates quite rapidly • In eggs but not sperm, so shows maternal inheritance The human Y chromosome • The mammalian X and Y chromosomes evolved from a pair of autosomes • The human Y has a block of material that transposed (moved) from the X since the divergence of chimps and humans Phylogeny based on haplotypes • If there has been no recombination, we can deduce the phylogeny of the haplotypes by parsimony Mutation (SNP) acgt Outgroup e.g. chimp acgt acga acca agca tgca Modern humans Dating the mutational events • The Y chromosome also has rapidly-mutating microsatellites (short tandemly repeated sequences such as CACACA….) • The older a mutation, the more diverse microsatellite alleles associated with it Haplogroup Ovchinnikov et al (2000) • Neanderthal mtDNA from Asia • Phylogenetic analysis of Neanderthal and modern human DNAs • Evidence for the Out-of-Africa model of modern human origins • Estimation of the age of the MRCA for modern humans and Neanderthals • Used phylogenies based on distance and parsimony Capelli et al (2003) • A Y-chromosome census of the British Isles • Compares Y chromosomes from regions of UK, with those in Denmark, Germany, Ireland, Norway • Different parts of UK have differing proportions of “indigenous” and “invading” Y chromosomes • Uses Y chromosome SNPs (which they call UEPs) and microsatellites • Capelli C, Redhead N, Abernethy JK, Gratrix F, Wilson JF, Moen T, Hervig T, Richards M, Stumpf MP, Underhill PA, Bradshaw P, Shaha A, Thomas MG, Bradman N, Goldstein DB. (2003) A Y chromosome census of the British Isles. Curr Biol. 13:979-84. Caramelli et al (2003) • Analysis of mtDNA from 24,000 year old anatomically modern (Cro-Magnon) humans • Evidence that Cro-Magnon types, not Neanderthals, are the ancestors of modern humans • Used multidimensional scaling to represent sequences as points in space, distance between them is a measure of genetic distance • Also used a simple measure of genetic distance Thomas et al (2000) • Lemba (“Black Jews”) are Bantu-speakers living in southern Africa • Claim Jewish origin based on stories and customs • Study compares their Y-chromosomes with other African and Jewish populations • Looked at distribution of haplotypes across populations, and proportions of haplotype sharing between populations • Also constructed genealogical tree of haplotypes The Exercise • Look at the 4 set papers • Highlight the questions we want you to investigate • You can decide what the other interesting questions are, and how they were answered • Organise yourself into group(s) to do this - each group should have at least one person who did the population genetics module • For each paper you should be able to understand how the main conclusions were reached and what were the important experimental results • Be ready to present your paper to the class at next meeting Additional papers (Not set this year) Brown et al (1998) • Studied the mtDNA of modern Native Americans 4 main types (A-D) • Minor 5th type (X) in Native Americans found also in Europeans but not Asians • So how did it get there? By crossing the Atlantic? • Used a network based on maximum parsimony to illustrate relationships of haplotypes, and coalescent analysis to estimate ages Helgason et al (2001) • mtDNA analysis to deduce the orgins of people in islands of North Atlantic (Iceland, Orkney, Scottish Western Isles, Skye, etc) • Relative contributions of Vikings from Scandinavia (about 1000 years ago) and Picts/Gaels from Britain/Ireland (more ancient) • Used r (rho) distance between samples as measure of the average number of base changes between a member of one population and a member of the other population Kayser et al (2000) • Study of the mutation rate and mechanism of Ychromosome microsatellites using father + son DNA • Important both for anthropological studies (dating the age of lineages) and in forensics • Used father-son DNA samples, a set of microsatellite polymorphisms, to calculate frequency and type of mutation • Also estimated the probability of true paternity