Molecular clocks and phylogenies
advertisement

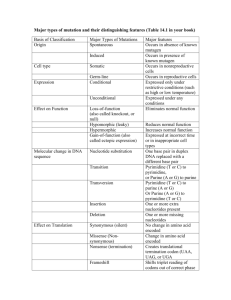
Genes and Evolution Molecular clocks and phylogenies Systematics A definition of taxonomy Phylogenies Characters and traits Homology and homoplasty Simple Phylogenies Molecular evolution Substitutions Synonymous and nonsynonymous Selection and neutral theory Molecular phylogenies Molecular clocks Adam Price Defining Systematics and Taxonomy Systematics- the study of the diversity of organisms Taxonomy- the science of classification of organisms taxis = Greek to arrange, classify Why? Explains evolutionary relationships Intrinsically interested Underpins an understanding of biology- e.g. ecology, conservation Important in applications to human life Phylogeny- the history of descent of a group of organisms from a common ancestor from Greek- phylon = tribe, race genesis = source Conventionally represented by a phylogeneic tree Monkey Gorilla Chimpanzee Human Ferns Conifers Peas Rice Phylogenic trees Phylogentic trees are based on comparison of traits individuals with common traits are placed together Character = a feature of the organisms (e.g. flower colour, height) Trait = one form of a character (blue flower colour, short height) Traits inherited from a common ancestor are termed homologous Traits that differs from the ancestor are termed derived Phylogenies are trees that best explain the distribution of homologous and derived traits in the organisms studied (the focal group). Scoring traits to make a simple phylogeny Taxon Hagfish Perch Salamander Lizard Crocodile Pigeon Mouse Chimpanzee Jaws Lungs + + + + + + + + + + + + + Claws or nails + + + + + Derived trait Feathers Fur + - + + Mammary glands + + Four-chambered heart + + + + A simple phylogeny Hagfish Perch Jaws Salamander Lungs Lizard Claws or nails Crocodile Feathers Four-chambered heart Pigeon Mouse Fur, mammary glands Chimpanzee Relative evolutionary time Ancient events Recent events Phylogenetic trees Outgroup Paraphyletic Polyphyletic Monophyletic Monophyletic taxa include all descendants of a common ancestor Paraphyletic taxa include some, but not all, descendants of a common ancestor Polyphyletic taxa includes members with more than one recent common ancestor Outgroup a lineage closely related to the focal group The problem of homoplastic traits traits that appear similar but are not related through ancestry Convergent evolution- independent evolution of similar traits due to similar selection pressure (e.g. wings in birds and bats) Parallel evolution- independent evolution of common traits in organisms sharing distant relatives (e.g. patterns of butterfly wings). Evolutionary reversals- the loss of a derived trait (e.g. limbs of snakes, teeth of frogs). Traits used in phylogenetics Morphology and developmental the importance of fossils Molecular Protein sequences Genetic markers DNA sequences The advantages of molecular traits 1/ They directly reflect the underlying process of evolution- changes in the hereditary material 2/ There are a vast number of potential traits 3/ They can detect difference between very closely related organism (even those that show no phenotypic difference) 4/ They are not effected by the environment (unlike some morphological traits) 5/ Since mutations generally occur as random events with specific probabilities, the number of mutations can be used to calibrate evolutionary time (molecular clocks) Disadvantages ? Transitions and Transversions in nucleotide substitutions Transitions replace a purine base with the other purine base, or a pyrimidine base with the other pyrimidine base Pyrimidines Purines TC CT AG G A Transversions replace a purine with a pyrimidine or vice versa A G Transition Transversion TA TG CA C G AT A C GT G C C Transition mutations are about 2 x more common than transversions T Transition Mutation in Coding vs Noncoding DNA A substantial part of the DNA of eukaryotes is noncoding- introns, repetitive sequences, pseudogenes Mutations in noncoding DNA do not generally effect phenotype and therefore are not subjected to selection Some mutations in coding regions do not change amino acid sequence because of the degenerate codon system Some mutations in coding regions change amino acid sequence to a similar type of amino acid, therefore having little or no effect on protein function Substitutions in Coding regions Synonymous vs nonsynonymous substitutions Synonymous substitutions are those that do not change the amino acid that is specified by the gene CUU ----> CUC = Leucine -----> Leucine Nonsynonymous substitutions are those that change the amino acid chain specified There are various degrees of nonsynonymous mutations depending on there effect on protein function. When the mutation changes the amino acid to a similar type then function may be little effected. Some amino acids of proteins are more important than othersactive sites for example. CUU ----> AUU = Leucine -----> Isoleucine Substitutions in Coding regions Miss-sense substitutions Miss-sense substitutions are those that prematurely terminate the gene UAU ----> UAG = Tyrosine -----> Stop UUA ----> UAA = Leucine -----> Stop Generally rare since nearly always involved change in protein activity Codon usage Second Letter U First Letter C A G U UUU UUC UUA UUG CUU CUC CUA CUG AUU AUC AUA AUG GUU GUC GUA GUG PhenylalanineF LeucineL LeucineL IsoleucineI MethionineM ValineV C UCU UCC UCA UCG CCU CCC CCA CCG ACU ACC ACA ACG GCU GCC GCA GCG SerineS ProlineP ThreonineT AlanineA A UAU UAC UAA UAG CAU CAC CAA CAG AAU AAC AAA AAG GAU GAC GAA GAG TyrosineY Stop HistidineH GlutamineQ AsparagineN LysineK Aspatic acidD Glutamic acidE G UGU UGC UGA UGG CGU CGC CGA CGG AGU AGC AGA AGG GGU GGC GGA GGG CystineC Stop TryptophanW ArginineR SerineS ArginineR GlycineG to ne 3 10 In su lin m yo gl ob in al bu in m te in ap r l eu ol ip ki op n ro 1 te in in Ate 1 rfe ro n B1 re la xi n hi s Substitutions per site per 1000,000,000 years Synonymous mutations are more commonly fixed in evolution 12 Synonymous mutations Nonsynonymous mutations 8 6 4 2 0 Substitutions per site per 1000,000,000 years 7 6 5 4 3 2 1 0 8 9 10 Animal mtDNA Pseudogenes Introns Fourfold degenerate sites Twofold degenerate sites Non-degenerate sites Downstream regions Upstream regions Different types of sequence evolve at different rates Comparing amino acid sequences Sequence 1 Sequence 2 leu arg phe cys ser ser arg Sequence 1 Sequence 2 leu arg phe cys ser ser arg Sequence 1 Sequence 2 Sequence 3 Sequence 4 Sequence 5 Sequence 6 leu leu leu leu leu leu leu phe cys ser ser arg leu gap phe cys ser ser arg arg gap gap arg arg arg phe cys ser ser phe cys ser phe phe cys ser phe ile cys ser ser ile cys ala ser phe cys ile ser arg arg arg arg arg arg Comparing amino acid sequences Sequence 1 Sequence 2 Sequence 3 Sequence 4 Sequence 5 Sequence 6 leu leu leu leu leu leu arg gap gap arg arg arg phe cys ser ser phe cys ser phe phe cys ser phe ile cys ser ser ile cys ala ser phe cys ile ser arg arg arg arg arg arg Sequence Number 1 1 Similarities 2 3 4 5 6 2 2 1 2 1 0 3 4 3 3 4 3 1 2 2 5 3 5 7 4 6 4 4 5 5 3 3 6 6 6 4 4 5 2 5 Differences Cytochrome C- a highly conserved gene Acidic side chains Hydrophobic side chains Basic side chains Human/chimp Rhesus monkey Horse Donkey Cow Dog Rabbit Gray whale Grey kangaroo G G G G G G G G G D D D D D D D D D V V V V V V V V V E E E E E E E E E K K K K K K K K K G G G G G G G G G K K K K K K K K K K K K K K K K K K I I I I I I I I I F F F F F F F F F I I V V V V V V V M M Q Q Q Q Q Q Q K K K K K K K K K C C C C C C C C C S S A A A A A A A Q Q Q Q Q Q Q Q Q C C C C C C C C C H H H H H H H H H T T T T T T T T T V V V V V V V V V E E E E E E E E E K K K K K K K K K G G G G G G G G G G G G G G G G G G K K K K K K K K K H H H H H H H H H K K K K K K K K K T T T T T T T T T G G G G G G G G G P P P P P P P P P N N N N N N N N N L L L L L L L L L G G G G G G G G G L L L L L L L L L F F F F F F F F F G G G G G G G G G R R R R R R R R R K K K K K K K K K T T T T T T T T T G G G G G G G G G Q Q Q Q Q Q Q Q Q A A A A A A A A A P P P P P P V V P G G G G G G G G G Y Y F F F F F F F S S T S S S S S T Y Y Y Y Y Y Y Y Y Chicken Pigeon Duck Turtle Snake Frog Tuna Dogfish G G G G G G G G D D D D D D D D I I V V V V V V E E E E E E A E K K K K K K K K G G G G G G G G K K K K K K K K K K K K K K K K I I I I I I T V F F F F F F F F V V V V T V V V Q Q Q Q M Q Q Q K K K K K K K K C C C C C C C C S S S A S A A A Q Q Q Q Q Q Q Q C C C C C C C C H H H H H H H H T T T T T T T T V V V V V C V V E E E E E E E E K K K K K K N N G G G G G G G G G G G G G G G G K K K K K K K K H H H H H H H H K K K K K K K K T T T T T V V T G G G G G G G G P P P P P P P P N N N N N N N N L H G L H G L H G L N G L H G L Y G L W G L S G L L L L L L L L F F F I F I F F G G G G G G G G R R R R R R R R K K K K K K K K T T T T T T T T G G G G G G G G Q Q Q Q Q Q Q Q A A A A A A A A E E E E V A E Q G G G G G G G G F F F F Y F Y F S S S S S S S S Y Y Y Y Y Y Y Y Samia moth G Hornworm moth G Screw worm fly G Fruit fly G Bakers yeast G Candida k rusei (yeast) G Neursopora (mold) G Wheat G Sunflower G Mung bean G Rice G Sesame G N N D D S S D N D D N D A A V V A A S P P S P V E D E E K K K D T K K K N N K K K K K A T S A S G G G G G G G G G G G G K K K K A A A A A E E E K K K K T T N K K K K K I I I L L L L I I I I I F F F F F F F F F F F F V V V V K K K K K K K K Q Q Q Q T T T T T T T T R R R R R R R K K K K K C C C C C C C C C C C C A A A A E A A A A A A A Q Q Q Q L E E Q Q Q Q Q C C C C C C C C C C C C H H H H H H H H H H H H T T T T T T V V V V V V E V V V V V E E E E E E A A A A K A E E E E E A K K K K G G G G G G N G G G G G G G G G G G L A A A A A K H K H K H K H P H P H T Q H H H H H K K K K K K K K K K K K V V V V V V I Q Q Q Q Q G G G G G G G G G G G G P P P P P P P P P P P P N N N N N N A N N N N N L L L L L L L L L L L L F F L L I I L L L L L L Y F F I F F F F F F F F G G G G G G G G G G G G R R R R R R R R R R R R K K K K H H K Q Q Q Q Q T T T T S S T S S S S S G G G G G G G G G G G G Q Q Q Q Q Q Q S T T T T A A A A A A A T T T T T P P A A P P D A A A P P G G G G G G G G G G G G F F F F Y Y Y Y Y Y Y Y S S A A S S A S S S S S Y Y Y Y Y Y Y Y Y Y Y Y T T T T T H H H H H H H H N H H H H H H H H N N N N G G G G G G G G G G G G Amino acid substitutions (per 100 residues) in cytochrome c A molecular clock for Cytochrome c 60 Angiosperms vs animals 50 Insects vs vertebrates 40 Yeast vs mould Mammals vs reptiles 30 Birds vs reptiles 20 Fish vs land vertebrates Amphibians vs birds and mammals 10 Birds vs mammals 0 0 200 400 600 800 1000 1200 Time since divergence (millions of years) The Neutral Theory of molecular evolution Most mutations are either selectively neutral or nearly so. Thus, the genetic variation within species results from random genetic drift Consider population of size N with a neutral mutation rate at a locus of mutations per gamete per generation No. of new mutations = x 2N Probability of fixation by genetic drift = frequency, p = 1/2N Number of new mutations per generation that are likely to become fixed by genetic drift = no. of mutations x probability of fixation = Molecular clocks The rate of fixation of neutral mutations is equal to the neutral mutation rate Thus, sequences diverge in evolution at a constant rate Thus, the divergence between two sequences can be used to say when the two organisms diverged from each other But remember •Not all mutations are neutral •Not all loci change at the same rate •Transitions are more common than transversions •Rates are strictly based on generations (not years), and reproductive rates vary between species Therefore, all molecular clocks need calibrating Calibration of -globin molecular clock Divergence of humans from other species based on -globin molecular clock calibrated on fossil evidence of divergence 600 Shark from cows 500 Frog 400 Chicken Alligator 300 200 Cow Quoll Million years ago Carp 100 Baboon 0 Species Divergence based on molecular clock Divergence based on fossil record