Algorithms for Nearest Neighbor Search
advertisement

Algorithms for Nearest Neighbor
Search
Piotr Indyk
MIT
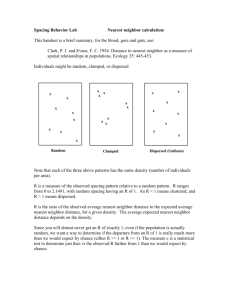
Nearest Neighbor Search
• Given: a set P of n points in Rd
• Goal: a data structure, which given a query
point q, finds the nearest neighbor p of q
in P
p
q
Outline of this talk
• Variants
• Motivation
• Main memory algorithms:
– quadtrees
– kd-trees
– Locality Sensitive Hashing
• Secondary storage algorithms:
– R-tree (and its variants)
– VA-file
Variants of nearest neighbor
• Near neighbor (range search): find one/all
points in P within distance r from q
• Spatial join: given two sets P,Q, find all
pairs p in P, q in Q, such that p is within
distance r from q
• Approximate near neighbor: find one/all
points p’ in P, whose distance to q is at
most (1+e) times the distance from q to its
nearest neighbor
Motivation
Depends on the value of d:
• low d: graphics, vision, GIS, etc
• high d:
– similarity search in databases (text, images etc)
– finding pairs of similar objects (e.g., copyright
violation detection)
– useful subroutine for clustering
Algorithms
• Main memory (Computational Geometry)
– linear scan
– tree-based:
• quadtree
• kd-tree
– hashing-based: Locality-Sensitive Hashing
• Secondary storage (Databases)
– R-tree (and numerous variants)
– Vector Approximation File (VA-file)
Quadtree
• Simplest spatial structure on Earth !
Quadtree ctd.
• Split the space into 2d equal subsquares
• Repeat until done:
– only one pixel left
– only one point left
– only a few points left
• Variants:
– split only one dimension at a time
– k-d-trees (in a moment)
Range search
• Near neighbor (range search):
– put the root on the stack
– repeat
• pop the next node T from the stack
• for each child C of T:
– if C is a leaf, examine point(s) in C
– if C intersects with the ball of radius r around q, add C to
the stack
Near neighbor ctd
Nearest neighbor
• Start range search with r =
• Whenever a point is found, update r
• Only investigate nodes with respect to
current r
Quadtree ctd.
• Simple data structure
• Versatile, easy to implement
• So why doesn’t this talk end here ?
– Empty spaces: if the points form sparse clouds,
it takes a while to reach them
– Space exponential in dimension
– Time exponential in dimension, e.g., points on
the hypercube
Space issues: example
K-d-trees [Bentley’75]
• Main ideas:
– only one-dimensional splits
– instead of splitting in the middle, choose the
split “carefully” (many variations)
– near(est) neighbor queries: as for quadtrees
• Advantages:
– no (or less) empty spaces
– only linear space
• Exponential query time still possible
Exponential query time
• What does it mean exactly ?
– Unless we do something really stupid, query time is at
most dn
– Therefore, the actual query time is
Min[ dn, exponential(d) ]
• This is still quite bad though, when the dimension
is around 20-30
• Unfortunately, it seems inevitable (both in theory
and practice)
Approximate nearest neighbor
• Can do it using (augmented) k-d trees, by
interrupting search earlier [Arya et al’94]
• Still exponential time (in the worst case)!
• Try a different approach:
– for exact queries, we can use binary search
trees or hashing
– can we adapt hashing to nearest neighbor
search ?
Locality-Sensitive Hashing
[Indyk-Motwani’98]
• Hash functions are locality-sensitive, if, for
a random hash random function h, for any
pair of points p,q we have:
– Pr[h(p)=h(q)] is “high” if p is “close” to q
– Pr[h(p)=h(q)] is “low” if p is”far” from q
Do such functions exist ?
• Consider the hypercube, i.e.,
– points from {0,1}d
– Hamming distance D(p,q)= # positions on
which p and q differ
• Define hash function h by choosing a set I
of k random coordinates, and setting
h(p) = projection of p on I
Example
• Take
– d=10, p=0101110010
– k=2, I={2,5}
• Then h(p)=11
h’s are locality-sensitive
• Pr[h(p)=h(q)]=(1-D(p,q)/d)k
• We can vary the probability by changing k
Pr
k=1
distance
Pr
k=2
distance
How can we use LSH ?
• Choose several h1..hl
• Initialize a hash array for each hi
• Store each point p in the bucket hi(p) of the
i-th hash array, i=1...l
• In order to answer query q
– for each i=1..l, retrieve points in a bucket hi(q)
– return the closest point found
What does this algorithm do ?
• By proper choice of parameters k and l, we can
make, for any p, the probability that
hi(p)=hi(q) for some i
look like this:
• Can control:
– Position of the slope
– How steep it is
distance
The LSH algorithm
• Therefore, we can solve (approximately) the near
neighbor problem with given parameter r
• Worst-case analysis guarantees dn1/(1+e) query time
• Practical evaluation indicates much better behavior
[GIM’99,HGI’00,Buh’00,BT’00]
• Drawbacks:
• works best for Hamming distance (although can be generalized
to Euclidean space)
• requires radius r to be fixed in advance
Secondary storage
• Seek time same as time needed to transfer
hundreds of KBs
• Grouping the data is crucial
• Different approach required:
– in main memory, any reduction in the number
of inspected points was good
– on disk, this is not the case !
Disk-based algorithms
• R-tree [Guttman’84]
– departing point for many variations
– over 600 citations ! (according to CiteSeer)
– “optimistic” approach: try to answer queries in
logarithmic time
• Vector Approximation File [WSB’98]
– “pessimistic” approach: if we need to scan the whole
data set, we better do it fast
• LSH works also on disk
R-tree
• “Bottom-up” approach (k-d-tree was “topdown”) :
– Start with a set of points/rectangles
– Partition the set into groups of small cardinality
– For each group, find minimum rectangle
containing objects from this group
– Repeat
R-tree ctd.
R-tree ctd.
• Advantages:
– Supports near(est) neighbor search (similar as
before)
– Works for points and rectangles
– Avoids empty spaces
– Many variants: X-tree, SS-tree, SR-tree etc
– Works well for low dimensions
• Not so great for high dimensions
VA-file [Weber, Schek, Blott’98]
• Approach:
– In high-dimensional spaces, all tree-based
indexing structures examine large fraction of
leaves
– If we need to visit so many nodes anyway, it is
better to scan the whole data set and avoid
performing seeks altogether
– 1 seek = transfer of few hundred KB
VA-file ctd.
• Natural question: how to speed-up linear
scan ?
• Answer: use approximation
– Use only i bits per dimension (and speed-up the
scan by a factor of 32/i)
– Identify all points which could be returned as
an answer
– Verify the points using original data set
Time to sum up
• “Curse of dimensionality” is indeed a curse
• In main memory, we can perform sublinear-time
search using trees or hashing
• In secondary storage, linear scan is pretty much all
we can do (for high dim)
• Personal thought: if linear search is all we can do,
we are not doing too well….
• Maybe it is time to buy a few GB of RAM
• ..but at the end everything depends on your data set
Resources
• Surveys:
– Berchtold & Keim:
– http://www.informatik.unihalle.de/~keim/PS/ICDE00.pdf
– Theodoridis:
– http://dias.cti.gr/~ytheod/research/ADBIS/handouts.pdf
– Agarwal et al (range searching):
– http://www.cs.duke.edu/~pankaj/papers.html
Resources
• Source code:
http://dias.cti.gr/~ytheod/research/indexing/
http://www.cs.sunysb.edu/~algorith/major_section/1.6.shtml
• References: see surveys plus very recent
– [Buh’00,BT’00]: J. Buhler et al:
http://www.cs.washington.edu/homes/jbuhler/
– [HGI’00]: Haveliwala et al:
http://theory.lcs.mit.edu/~indyk/webdb.ps
Contact
• If you have any question, feel free to e-mail
me at indyk@theory.lcs.mit.edu
• Thank you !