Analytical Loop Closure & Applications
advertisement

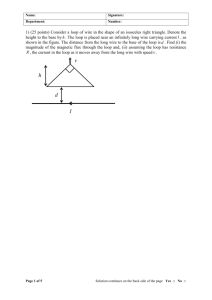
Geometrical RING Optimization Evangelos Coutsias Dept of Mathematics and Statistics, Univ. of New Mexico Jointly with Chaok Seok, Matthew Jacobson, and Ken Dill Dept of Pharmaceutical Chemistry, UCSF Michael Wester Office of Biocomputing, UNM Abstract In previous work, we considered the problem of loop closure, i.e., of finding the ensemble of possible backbone structures of a chain molecule that are consistent geometrically with preceding and following parts of the chain whose structures are given. We provided a simple intuitive view and derivation of a 16th degree polynomial equation for the case in which the six torsion angles used for the closure belong in three coterminal pairs. Our work generalized previous results on analytical loop closure as our torsion angles need not be consecutive, and any rigid intervening segments are allowed between the free torsions. We combined the new scheme with an existing loop construction algorithm to sample protein loops longer than three residues and used it to implement a set of local moves for Monte Carlo minimization. Here we present an application to the sampling of S2-bridged 9-peptide loops and discuss the implementation of the local moves as a Metropolis Monte Carlo scheme for the uniform sampling of conformational space. With the base Cn 1 Cn 1 and the lengths of the two peptide virtual bonds fixed, the vertex C n is constrained to lie on a circle. Tripeptide Loop Closure N Cn C C' Cn 1 Cn 1 Bond vectors fixed in space Fixed distance o d min 4.7 The triangle formed by three consecutive Ca atoms: Given the span, d, there are constraints on the orientation of the middle Cb, the side chain and the two coterminal peptide units about the virtual bonds between the Ca (green circles). o d max 7.3 Designing a 9-peptide ring 111.6 o 16.63o 19.13o 90 o Given the span, the two consecutive peptide units are correlated 90 o 2 1 This extends to the orientation of Cb 111 o A bimodal example 111 o 110 10 o Thetaperturbations are not enough o 111 o 19 5 o o 111 o 19 5 o o 1r69: Res 9-19 alternative backbone configurations Representation of Loop Structures In the original frame In the new frame New View of Loop Closure Old View New View 6 rotations / 6 constraints 3 rotations / 3 constraints x2 x1 x3 Follower R4 1 G2 a2 Crank R1 a1 G3 a3 1 a4 z4 1 s1 s4 Two-revolute, two-spheric-pair mechanism z1 x c O y z The 4-bar spherical linkage F ( ; , , , ) y Transfer Function for concerted rotations s1 p r s2 d R r R p z x t1 t2 R t 2 1 cos ( R p ) cos cos tan 2 2 R t1 R t R t 1 2 1 z y R1 R2 1 2 x L1 L2 4 3 A complete cycle through the allowed values for (dihedral (R1,R2) -(L1,R1) )and y (dihedral (R1,R2)-(L2,R2)) 3 2 1 0 -1 t -2 -3 2 4 6 8 10 flywheel equations 12 3 y1, y2 2.5 2 .35*p y2 0 1.5 1 r1=.81=r2 0.5 0 -3.5 -3 -2.5 -2 -1.5 y1 -1 -0.5 0 0.5 Derivation of a 16th Degree Polynomial for the 6-angle Loop Closure 2 ri-1 ·ri = cos i gives Pi(ui-1, ui) =j,k=0 pjk ui-1j uik , where ui = tan(i/2). Using the method of resultants, the three biquadratic equations P1(u3, u1), P2(u1, u2), and P3(u2, u3) are reduced to a polynomial in u3, r 16 R16(u3) = rj u3j j=0 r1 2 Method of resultants gives an equivalent 16th degree polynomial for a single variable zi , (i 1,2,3) Numerical evidence that at most 8 real solutions exist. Must be related to parameter values: the similar problem of the 6R linkage in a multijointed robot arm is known to possess 16 solutions for certain ranges of parameter values (Wampler and Morgan ’87; Lee and Liang ‘’89). The Minkowski sum of three squares, of side a, b, c resp. Here a=2x, b=2y, c=2z are the sizes of three scaled Newton polytopes for the three biquadratics 0,0, a c 0, b c,0 a 2x b 2y a b, b c, c a c 2z a b,0,0 There are at most 16 solutions: from first principles V a, b, c a b b c c a 2abc a 2 b c b 2 c a c 2 a b 16 xyz By the Bernstein-Kushnirenko-Khovanski theorem the total number of isolated solutions cannot exceed the mixed volume of the Minkowski sum of the Newton Polytopes of the consitutive polynomial components. That is, the number 16 is generic for this problem. Methods of determining all zeros: (1) carry out resultant elimination; derive univariate polynomial of degree 16 solve using Sturm chains and deflation (2) carry out resultant elimination but convert matrix polynomial to a generalized eigenproblem of size 24 (3) work directly with trigonometric version; use geometry to define feasible intervals and exhaustively search. It is important to allow flexibility in some degrees of freedom Loop Closure Algorithm 1. Polynomial coefficients are determined in terms of the geometric parameters on the right. 2. u3 = tan(3/2) is obtained by solving the polynomial equation. 3, 1, and 2 follow. 3. Positions of the all atoms are determined by transforming to the original frame. 1 2 3 General Chain Loop Closure 7-Angle Loop Closure The continuous move used in Monte Carlo energy minimization 2b 2 4 2a 3 1 1a 1 2 1b 1 4a 4 3' 4b 4 5a 5 5b 5 5 The continuous move: given a state assume D2b, D4a fixed, but D3 variable tau2sigma4 determined by D3 (1) tau1sigma2, tau4sigma5 trivial (2) alpha1, alpha5 variable but depend only on vertices as do lengths (lengths 1-2, 1-5, 4-5 are fixed) Given these sigma1tau1, sigma5tau5 known (sigma1tau5 given) (3) Dihedral (2-1-5-4) fixes remainder: alpha2, alpha4 determined (sigma2tau2, sigma4tau4 known) Longer Loop Closure in Combination with an Existing Loop Construction Method Analytical closure of the two arms of a loop in the middle Coutsias, Seok, Jacobson and Dill, J Comp Chem 25(4), 510 (2004) Jacobson, et al, Proteins, 2004. Canutescu and Dunbrack, Protein Science, 12, 963 (2003). Refinement of 8 residue loop (84-91) of turkey egg white lysozyme Native structure (red) and initial structure (blue) Baysal, C. and Meirovitch, H., J. Phys. Chem. A, 1997, 101, 2185 pep virtual bond 3-pep bridge design triangle C 9-pep ring cysteine bridge 1 2 Modeling R. Larson’s 9-peptide B A 3 Designing a 9-peptide ring In designing a 9-peptide ring, the known parameters of 2-pep bridges (and those of the S2 bridge, if present) are incorporated in the choice of the foundation triangle, with vertices A,B,C (3 DOF) lmin l1 lmax C l1 l2 B l3 A C d2 d1 l1 l3 l2 B A d3 peptide virtual bond (3 dof for placement)x3=9 2-pep virtual bond (at most 8 solutions) design triangle sides (3 dof ) 8-2-4 4-2-4 Cyclic 9-peptide backbone design 4-6-2 4-2-2 Numbers denote alternative loop closure solutions at each side of the brace triangle Disulfide Loop Closure • Start at C of the “last” Cysteine residue • The dihedral angle 2 is a free variable: vary continuously to get all possible conformations. • Fix the bonds: N 9 C ,9 and C ,1 C1 • Note that a move rooted at the “first” Cysteine must not fix N1 C ,1 but rather C ,1 C ,1 Disulfide Bridge Loop Closure PEP25: CLLRMKSAC 4 8 16 24 32 40 9998 2319 -227.911 2.89 -234.104 2.66 9999 0 -214.445 2.07 -234.104 2.66 10000 0 -216.965 2.79 -234.104 2.66 ---------------------------------------------------------n_trial, n_accept, ratio: 10000 2319 0.2319000 ---------------------------------------------------------min E = -234.104 rmsd = 2.664 num saved pdb = 40 Number of total energy evaluations = 1788596 ---------------------------------------------------------Total User Time = 10004.895 sec 0 dy 2 hr 46 min 44.895 sec # Final time: Apr 10 22:24:29 2004 0.6 120 N 0 1.4 273 N 0 0.4 90 N 0 RMSD vs Time 0.25 0.20 RMSD(nm) 0.15 0.10 0.05 0.00 400 800 1200 1600 2000 2400 2800 3200 time (ps) CLLRMRSIC MD Calculation using GROMAX with explicit water by Ilya Chorny, UCSF 3600 4000